Xenoanthostomella Mapook & K.D. Hyde, in Hyde et al., Fungal Diversity 100: 235 (2020)
Index Fungorum number: IF 556907; MycoBank number: MB 556907; Facesoffungi number: FoF 06795
Etymology – Xeno = ξένος in Greek, distinct, Anthostomella = anthostomella-like.
Saprobic on dead stems. Sexual morph: Ascomata immersed beneath clypeus, unilocular, globose to obpyriform, coriaceous, solitary to scattered. Clypeus extending outwards around the ascomata, thicker around the papilla. Ostiole central, papillate. Peridium comprising several layers, inner layers comprising hyaline to brown cells of textura epidermoidea, outer layers comprising brown cells of textura intricata at the side with textura angularis at the upper part. Paraphyses cylindrical, septate with small guttules, hyaline, branching, and embedded in a gelatinous matrix. Asci 8-spored, unitunicate, cylindrical to broadly filiform, pedicellate, straight or slightly curved, with an inconspicuous apical ring. Ascospores 1-seriate, hyaline when immature, becoming brown to dark olivaceous-brown when mature, unicellular, inequilateral to broadly fusiform, tapering towards narrow ends, verruculose when immature, with 1–2 large guttules when mature. Asexual morph: Undetermined.
Type species – Xenoanthostomella chromolaenae Mapook & K.D. Hyde.
Notes – A phylogenetic analysis based on a combined data- set of ITS, LSU, RPB2, TUB2, SSU and TEF1-α sequence data showed that Xenoanthostomella chromolaenae clusters with Idriella lunata, Gyrothrix ramosa and Gyrothrix inops (Fig. 153). However, we could not compare the morphological characteristics of these genera as Xenoanthostomella is found as a sexual morph in nature and asexual morph did not form in culture, while Idriella, Gyrothrix and Circinotrichum were found only as asexual morphs in nature. Therefore, Xenoanthostomella is described here as a new genus based on phylogeny, together with a comparison of the ITS (+5.8S) gene region of X. chromolaenae and Gyrothrix ramosa reveals 13 base pair differences (2.56%) across 507 nucleotides.
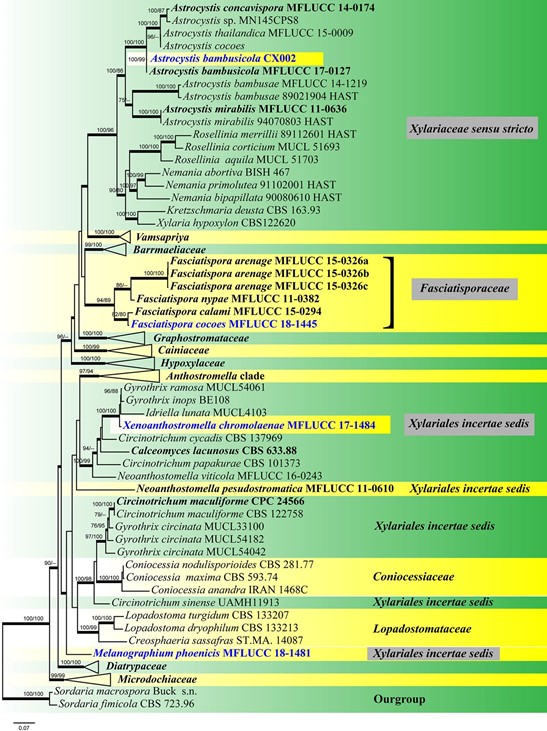
Fig. 1 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, RPB2, TUB2, SSU and TEF1-α sequence data representing the order Xylariales. Related strains are referred to as Hsieh et al. (2010), Daranagama et al. (2015), Liu et al. (2015), Becerra-Hernández et al. (2016), Maharachchikumbura et al. (2016), Dai et al. (2017), Hyde et al. (2017), Wendt et al. (2018), Voglmayr et al. (2018). Ninety-three strains are included in the combined analyses which comprise 6715 characters (828 characters for ITS, 885 characters for LSU, 1063 characters for RPB2, 1990 characters for TUB2, 1025 characters for SSU, 924 characters for TEF1-α) including gaps. Sordaaria fimicola (CBS 723.96) and Sordaria macrospora (Buck s.n.) are used as the outgroup taxa. Single gene analyses were also performed to compare the topology and clade stability with combined gene analyses. The tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RAxML tree with a final likelihood value of -69169.901135. The matrix had 3540 distinct alignment patterns, with 60.93% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241739, C = 0.259560, G = 0.259228, T = 0.239474; substitution rates AC = 1.286285, AG = 3.197641, AT = 1.369896, CG = 1.063194, CT = 5.517353, GT = 1.000000; gamma distribution shape parameter α = 0.718586. Bootstrap values for maximum likelihood (ML) and maximum parsimony equal to or greater than 75%, and clade credibility values greater than 0.95 are indicated in bold branches. The new isolates are indicated in bold and blue, and the type strains are indicated in black bold.
Species
