Valsaria rudis (P. Karst. & Har.) Theiss. & Syd. ex Petr. & Syd., (1923)
MycoBank number: MB 277020; Index Fungorum number: IF 277020; Facesoffungi number: FoF 00611; Fig. 17
≡ Dothidea rudis P. Karst. & Har. 1889
Saprobic on dead branch of Quercus sp. Sexual morph: Stromata 1.2–1.5 mm high, 1.7–2.5 mm diam, pseudostromatic, erumpent from host surface, scattered or rarely gregarious, pustular at dehiscence, broadly conical or subglobose with flattened base, enclosed on top and/or at the sides by a thick pseudoparenchymatous black crust, 40–55 μm thick between adjacent stromata. Ectostroma forming inversely stellate structures of 3–5 greyish, or greenish to black tubercular segments, the tissue beneath thick pseudoparenchymatous crust, tissue at the stromatal base prosenchymatous, grey, mixed with bark cells. Ostiolar inconspicuous opening at the surface. Ostiolar necks 0.3–0.5 mm high, cylindrical or conical.Ascomata 0.4–0.6 mm high, 0.3–0.6 mm diam, arranged in valsoid configuration, 9–20 ascomatal structures per individual cluster, subglobose to conical, without ostiolar neck. Peridium 25–40 μm thick, comprising pale brown to dark cells of textura angularis. Hamathecium comprises numerous, apically free pseudoparaphyses 1–2.5 µm wide, dense, filamentous, tapering towards apex, aseptate, hyaline. Asci 70–120 × 7–15 μm (x̅ = 85–10 µm, n = 20), 8-spored, bitunicate, fissitunicate, cylindrical, short pedicel, apically rounded, containing an ocular chamber and a pulvinate ring. Ascospores 14–20 × 6–9 μm, (x̅ = 16.6–8.5 µm, n = 20), uni-seriate, ellipsoid, hyaline when immature, becoming pale brown to dark brown at maturity, 1-septate, sometimes constricted at the septum, rounded at the both ends, with two distinct guttules, surface finely warted to reticulate. Asexual morph: Coelomycetous, see Jaklitsch et al. (2015).
Material examined – Italy, Province of Forlì-Cesena [FC], Rocca delle Caminate – Predappio, dead and fallen branch of Quercus sp. (Fagaceae), 2 March 2017, Erio Camporesi, IT 3268 (MFLU 17-0730, HKAS 102333), ibid., 22 March 2017, IT3268a (MFLU 17-0842, HKAS 102345); living culture MFLUCC 18-0532.
GenBank numbers – ITS: OM614589, OM614590; LSU: OM616560, OM616561; rpb2: ***, tef1-α:***.
Notes – In the phylogenetic analysis of Valsariaceae, our strains MFLUCC 18-0532 and MFLU 17-0730 grouped with strains of Valsaria rudis (V3, CBS 139066, VQC, CBS 139065, V7 and V31) with 99% MLBS and 1.00 BYPP support (Fig. 18). Jaklitsch et al. (2015) designated a lectotype for Valsaria rudis (H, herb. Karsten 3713) from France based on morpho-molecular evidence and further examined a strain of V. rudis reported from Lazio, Italy (WU 33486; culture V3) on Quercus cerris trees. The known distribution of V. Rudis is suggested to be host-specific on Quercus species (Jaklitsch et al. 2015). The absence of ectostroma and the presence of cupulate ostiolar discs on the surface of the stroma are key morphological characters of V. rudis. Furthermore, MFLU17-0730 and MFLU 17-0842 share identical morphology to V. rudis (WU 33485) in Jaklitsch et al. (2015). This is the first report of V. rudis on Quercus sp. from the Emilia–Romagna region.

Figure 17 – Valsaria rudis (MFLU 17-0730, MFLU 17-0842). a–c. Appearance of ectostromata on a dead and fallen branch of Quercus sp. d. Longitudinal view of stroma. e. Peridium. f. Apically free pseudoparaphyses. g–j. Asci. (g; pulvinate ring is arrowed) k–n. Two-cellular ascospores. o. Germinating ascospore. p, q. Culture characteristics on PDA from surface (p) and reverse (q). Scale bars: a = 1 mm, b, d = 500 µm, c = 200 µm, j = 20 µm, e–i = 10 µm, k–o = 5 µm.
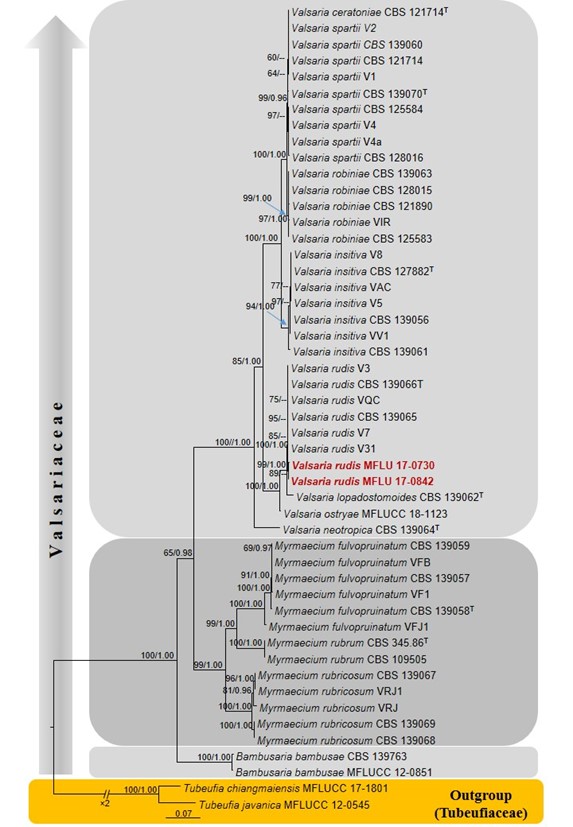
Figure 18 – Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, rpb2 and tef1-α sequenced data. Fifty strains were included in the combined sequence analyses, which comprised 3762 characters with gaps (LSU = 899, ITS = 552, rpb2 = 1183, tef1-α = 1129). Single gene analyses were also performed and topology and clade stability compared from combined gene analyses. Tubeufia chiangmaiensis (MFLUCC 17-1801) and T. javanica (MFLUCC 12-0545) were used as the outgroup taxa. Final ML optimization likelihood is -17441.380242. The matrix included 1217 distinct alignment patterns, with 8.60% undetermined characters or gaps. Estimated base frequencies were obtained as follows: A = 0.246660, C = 0.264801, G = 0.271013, T = 0.217526; substitution rates AC = 1.466946, AG = 3.413440, AT = 1.139395, CG = 0.993542, CT = 8.729450, GT = 1.0; gamma distribution. Bootstrap support values for ML (first set) equal to or greater than 70%, BYPP equal to or greater than 0.95 are given above or below the nodes. The strains from the current study are bold and the type strains are indicated with T.
