Spinulosphaeria Sivan., Trans. Br. mycol. Soc. 62(1): 5 (1974)
MycoBank number: MB 5149; Index Fungorum number: IF 5149; Facesoffungi number: FoF 10527; 2 morphological species (Species Fungorum 2020), 1 species with sequence data.
Type species – Spinulosphaeria thaxteri (Pat.) Sivan.
Notes– Spinulosphaeria, typified by Spinulosphaeria thaxteri, was established by Sivanesan (1974) and has tuberculate ascomata, clavate asci and subglobose to ellipsoid ascospores. The second species, Spinulosphaeria nuda is characterized by tuberculate ascomata and verrucose ascospores with mucilaginous sheath (Mugambi & Huhndorf 2010). Phylogenetic analysis of LSU sequence data of S. nuda reveals that Spinulosphaeria is related to Chaetosphaerellaceae and Nitschkiaceae (Mugambi & Huhndorf 2010). In this study, Spinulosphaeria is closely related to Crassochaeta and Chaetosphaerella within Chaetosphaerellaceae (Fig. 11).
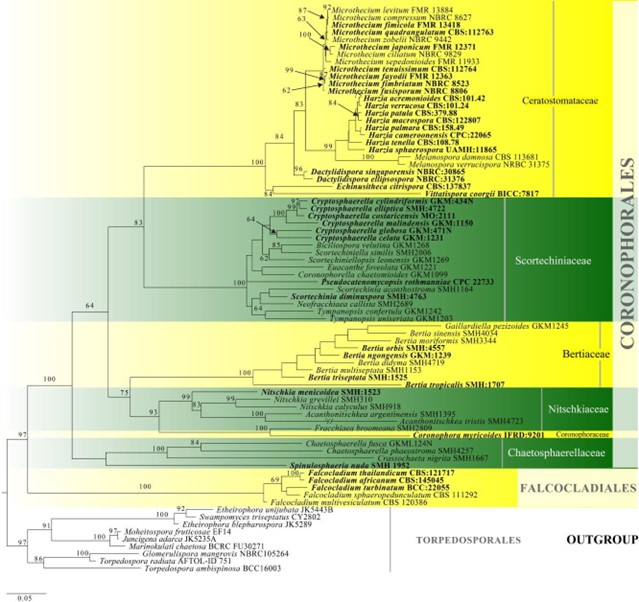
Figure 11 – Phylogram generated from maximum likelihood analysis based on combined LSU, tef1 and ITS sequence data for Coronophorales. Related sequences are referred to Hongsanan et al. (2017). Seventy-nine strains are included in the combined analyses which comprised 2211 characters (1044 characters for LSU, 627 characters for tef1, 540 characters for ITS) after alignment. Members of Torpedosporales are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. The best RaxML tree with a final likelihood value of -23354.525618 is presented. Estimated base frequencies were as follows: A = 0.228019, C = 0.288494, G = 0.281955, T = 0.201533; substitution rates AC = 1.194725, AG = 2.787601, AT = 1.703067, CG = 0.955075, CT = 3.781057, GT = 1.000000; gamma distribution shape parameter a = 0.736138. Bootstrap support values for ML greater than 50% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
Species
