Pseudophialocephala M.S. Calabon, E.B.G. Jones, K.D. Hyde, gen. nov.
MycoBank number: MB 559318; Index Fungorum number: IF 559318; Facesoffungi number: FoF 10602;
Etymology – Name reflects the morphological similarity to Phialocephala
Saprobic on decaying wood from terrestrial and aquatic habitats. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies on natural substrate effuse, white, brown, velvety. Mycelium partly superficial to immersed, composed of hyaline, branched, septate, constricted at the septa, guttulate hyphae, 1.5–3 μm wide. Conidiophore determinate, macronematous, scattered, solitary, composed of stipe and penicillate heads. Stipe thick-walled, erect, straight, or broadly curved, cylindrical, brown, and wide at base, paler and slightly tapering toward the apex, terminating in a penicillate head. Penicillate heads composed of 1–3 series of metulae terminating in groups of conidiogenous cells. Metulae thin-walled, light brown to subhyaline. Conidiogenous cells monophialidic, integrated, discrete, determinate, terminal, clustered at the apex of secondary metulae, hyaline, subhayaline to pale brown, thin-walled, cylindrical, with inconspicuous openings, with minute collarettes. Conidia um, acrogenous, aseptate to 1-septate, aggregated in slimy and white masses, solitary to catenate, hyaline, ellipsoidal, cylindrical, rounded at each end, smooth, thin walled, guttulate.
Type species – Pseudophialocephala humicola (S.C. Jong & E.E. Davis) M.S. Calabon, E.B.G. Jones, K.D. Hyde
Notes – Phialocephala sensu stricto, clade containing the type species P. dimorphospora, are saprotrophs mostly collected in temperate climates (Wijayawardene et al. 2020; Tanney and Seifert 2020). Species under Phialocephala s.s. include P. aylmerensis, P. biguttulata, P. botulispora, P. catenospora, P. cladophialophoroides, P. collarifera, P. dimorphospora, P. heterosperma, P. lagerbergii, P. lignicola, P. mallochii, P. nodosa, P. oblonga, and P. repens and clustered within Mollisiaceae. Phialocephala sensu lato is currently polyphyletic and it includes P. fluminis (Chaetothyriales), P. fusca (Ophiostomatales), P. hiberna (Mollisiaceae), P. humicola (Ophiostomatales, Sordariales), P. scopiformis (Mollisiaceae), P. virens (Ophiostomatales), and P. xalapensis (Ophiostomatales)
(Grünig et al. 2002; Jacobs et al. 2003; Day et al. 2012; Tanney and Seifert 2020). Tanney and Miller (2017) suggested the placement of P. fusca and P. humicola in Chaetosphaeriales based on the BLASTn result of the ITS sequence data. In the present study, the combined LSU-ITS phylogenetic analyses confirm the placement of P. humicola in Chaetosphaeriaceae (Fig **). Phialocephala fusca (CBS 301.85), Chloridium lignicola (CBS 143.54) and C. pini (CPC 36627) formed an independent clade basal to the taxa of Chaetosphaeriaceae and distant to the clade shared by P. humicola and other taxa of Chloridium and Sporochisma. Since P. humicola was morphologically different from the generic description of Chloridium and Sporochisma, we introduce a novel genus Pseudophialocephala and transfer C. aquaticum, C. salinicola, and C. terricola to this genus as they cluster together with P. humicola. A novel Pseudophialocephala taxa, P. cuneata, collected form decaying wood in Thailand, was introduced based on morphology and phylogenetic analysis of LSU and ITS sequence data. Currently, three Pseudophialocephala species were saprobic from aquatic habitats: freshwater (P. aquatica) (Wei et al. 2018), marine (P. humicola, P. salinicola) (Dayarathne et al. 2020, this study), and in terrestrial habitats (P. cuneata, P. humicola, P. terricola, P. xalapensis) (Jong and Davis 1972; Kiyuna et al. 2012; Chunyu et al. 2013).

Figure 33 – Phylogenetic tree generated by ML analysis of combined LSU and ITS data of Chaetosphaeriales and closely related taxa. The analyses included 141 strains and the tree was rooted with Tremella mesenterica (CBS 6973) and Trichosporon asahii (CBS 2479). Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of -23625.921520 is presented. The matrix had 843 distinct alignment patterns, with 22.02% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231971, C = 0.256425, G = 0.298236, T = 0.213368; substitution rates AC = 1.373661, AG = 2.038871, AT = 1.670986, CG = 0.755571, CT = 5.839863, GT = 1.000000; gamma distribution shape parameter α = 0 0.347250. RAxML bootstrap support values ≥75% and Bayesian posterior probabilities values ≥0.95 (BYPP) are shown near the nodes. New isolates recovered in this are in red.

Figure 33 – Continued.
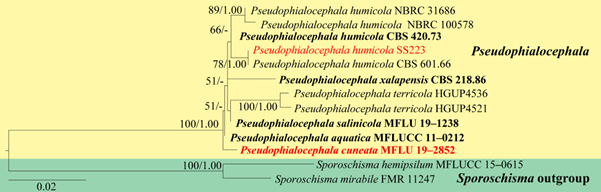
Figure 34 – Phylogenetic tree generated by ML analysis of combined LSU and ITS sequence data of Pseudophialocephala. The analyses included 13 strains and the tree was rooted with Sporoschisma hemipsilum (MFLUCC 15–0615) and Sporoschisma mirabile (FMR 11247). Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of -2920.924831 is presented. The matrix had 106 distinct alignment patterns, with 6.78% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231239, C = 0.266364, G = 0.302672, T = 0.199726; substitution rates AC = 2.586006, AG = 3.386907, AT = 1.676969, CG = 1.508214, CT = 6.123811, GT = 1.000000; gamma distribution shape parameter α = 0.225772. RAxML bootstrap support values ≥75% and Bayesian posterior probabilities values ≥0.95 (BYPP) are shown near the nodes. Ex–type/ ex–epitype strains are in bold. New isolates recovered in this are in red.
