Prosthemium betulinum Kunze, in Kunze & Schmidt, Mykologische Hefte (Leipzig) 1: 18 (1817)
Index Fungorum number: IF 223054; MycoBank number: MB 820836; Facesoffungi number: FoF 06080; Figs. 20, 21
Saprobic in terrestrial habitats. Sexual morph: Ascomata 879–979 × 284–401 μm (x̅ = 922.9 × 339.2 μm, n = 10), solitary, scattered, or in small groups, immersed, erumpent or depressed globose, medium to large, black, ostiolate. Ostiole papillate, opening via a minute slit or a small conical swelling in the bark, ostiolar canal filled with a tissue of hyaline small cells. Peridium 1–2 layered comprised of compressed cells of textura angularis, apex comprises comparatively larger cells than the base. Hamathecium of dense, cellular, filiform, broad, septate pseudoparaphyses, embedded in a gelatinous matrix. Asci 77–186 × 30–42 μm (x̅ = 140.1 × 36.4 μm, n = 10), 8-spored, bitunicate, fissitunicate, ellipsoid, broadly cylindrical to broadly cylindro-clavate, with a short, narrow, thick pedicel, rounded at the apex with a minute ocular chamber. Ascospores 52–73 × 14–16 μm (x̅ = 61.3 × 15.4 μm, n = 10), biseriate, partially overlapping, narrowly oblong with broadly to narrowly rounded ends, brown to golden brown, muriform, 5–8 transverse septa and 1–2 vertical septa in some cells, constricted at each septum, smooth to verrucose wall with a mucilaginous sheath. Asexual morph: Pycnidia 390–359 × 301–316 μm (x̅ = 302.3 × 310.1 μm, n = 10), globose to subglobose, brown to black. Conidiophores 10–14 × 4–6 μm (x̅ = 12.7 × 5.3 μm, n = 10), unbranched, hyaline, septate. Macroconidia brown, staurosporous, 49–95 μm diam with two main arms and 2 or 3 smaller arms. Main arms 22–33 × 14–19 μm (x̅ = 28.2 × 16.3 μm, n = 20), 4–5 transversely septate, with terminal cell hyaline to pale brown. Smaller arms continuous or 1–3 septate, hyaline to pale brown. Microconidia 4–5 × 2–3 μm (x̅ = 4.7 × 3.2 μm, n = 10), brown to dark brown, sometimes with 1–2 guttules, globose to ellipsoid, aseptate, thick-walled, smooth.
Culture characteristics: Colonies on MEA, 10–15 mm diam after 7 days at 16 ºC, margin regular, slightly raised, rough surface, entire edge, cottony or woolly, well-defined edges with no pigmentation of the agar.
Material examined: ITALY, Via Togliatti—Forlì (Province of Forlì-Cesena [FC])—Italy, dead land branch of Betula sp. (Betulaceae), 26 March 2018, E. Camporesi (MFLU 17-0975); HKAS 97499.
GenBank numbers: LSU: MK652791; SSU: MK652791; ITS: MK652789.
Notes: Isolate MFLUCC 17-2312 was recovered from a dead branch of Betula sp. from Forli-Cesena Province. The new isolate clusters with other Prosthemium betulinum strains with moderate bootstrap support (65% ML/1.00 PP) (Fig. 22). The ITS sequence similarity between the new isolate and other P. betulinum strains is 100% (Jeewon and Hyde 2016). Therefore, we introduce P. betulinum as a new record from Italy

Fig. 20 Prosthemium betulinum. (MFLU 17-0975, a new geographical record). a,b Appearance of ascomata on host surface. c Vertical section through the ascomata. d Peridium e–g Asci. h Asci showing ocular chamber i Ascospore in Indian ink showing sheath j-l Ascospores m Germinating ascospores n,o Culture characteristics on MEA (n: reverse view; o: above view) Scale bars: a = 1000 μm, b = 2000 μm, c = 400 μm, d = 40 μm, e–g = 100 μm, h–l = 20 μm, m = 50 μm
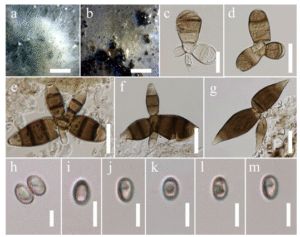
Fig. 21 Prosthemium betulinum. Asexual Morph (MFLU 17-0975, a new geographicalrecord). a,b Pycnidia on culture. c–g Macroconidia. h–m Microconidia. Scale bars: a = 1000 μm, b = 1000 μm, c = 15 μm, d–g = 20 μm, h–m = 5 μm
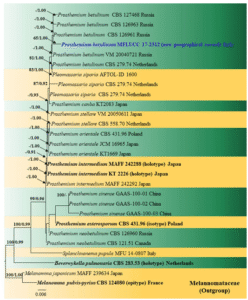
Fig. 22 Phylogram generated from maximum likelihood analysis based on a combined ITS, LSU and SSU sequence dataset retrieved from the GenBank. Melanomma pulvis-pyrius and M. japonicum are used as the out-group taxa. ML bootstrap values ≥ 50% are given as the first set of numbers and approximate likelihood-ratio test (aLRT) ≥ 0.90 values as the second set of numbers above the nodes. Voucher/strain numbers are given after the taxon names, the ones from type material are indicated in bold face. Newly generated sequence is indicated in blue. The bar length indicates the number of nucleotide substitutions per site
