Populomyces zwinianus Hern.-Restr., in Crous et al., Fungal Systematics and Evolution 7: 321 (2021)
MycoBank number: MB 838713; Index Fungorum number: IF 838713; Facesoffungi number: FoF 14116.
Etymology: Name refers to the school from where the sample was collected “Zwin College” (Oostburg, the Netherlands). This sample was collected during a Citizen Science project at the Westerdijk Fungal Biodiversity Institute.
Vegetative hyphae hyaline, septate, smooth, 1–2 µm wide. Conidiophores micronematous, mostly reduced to conidiogenous cells. Conidiogenous cells monophialidic, aggregated in groups, hyaline, smooth, cylindrical to lageniform, 5–16 × 2–3.5 µm, with a cylindrical collarette, 0.5–1.5 µm deep, 1–1.5 µm wide. Conidia aseptate, cylindrical, solitary, hyaline, guttulate, 9–12 × 2–2.5 µm.
Culture characteristics: After 2 wk at 24 °C on OA reaching 15 mm diam, membranous, saffron; sparse aerial mycelium, margin regular; reverse pale luteous. On MEA reaching 10 mm diam, elevated, with a radial disposition of the wrinkles, velvety, peach to flesh, margin slightly lobate; reverse saffron to apricot.
Typus: Netherlands, Zeeland Province, Yerseke, from soil, 11 Jun. 2019, W. Vercouteren, S. Meas & R. Verhije, NL19_076 (holotype CBS H-24737, ex-type strain CBS 147307 =NL1976004).
Notes: Populomyces is phylogenetically close to Calloria and Tricellula (Fig. 2). The cylindrical, aseptate conidia of Populomyces are easily distinguished from the stauroconidia of Tricellula (Seifert et al. 2011). Furthermore, Calloria is a polyphyletic genus with apothecial ascomata including species that are related to Cylindrocolla asexual morphs, characterised by polyblastic conidiogenous cells producing conidia in chains, thus distinct from the solitary conidia of Populomyces (Muntañola-Cvetkovic et al. 1997, Seifert et al. 2011).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Chalara sp. (GenBank KX034388.1; Identities = 463/510 (91 %), one gap (0 %)), Dothideomycetes sp. (strain CK1374, GenBank MH474014.1; Identities = 444/493 (90%), 16 gaps (3 %)), and Fungal sp. (strain LEG-103, GenBank MW201484.1; Identities = 446/496 (90 %), 15 gaps (3 %). Closest hits using the LSU sequence are Calloria urticae (strain G.M. 2016- 03-12.3, GenBank MT509570.1; Identities = 630/669 (94 %), four gaps (0 %)), Calloria urticae (strain MFLU 18-0697, GenBank MK591969.1; Identities = 824/824 (100 %), four gaps (0 %)), and Calloria urticae (strain MFLU 18-0696, GenBank MK591968.1; Identities = 630/669 (94 %), four gaps (0 %)) – also see Fig. 2.
Author: M. Hernández-Restrepo
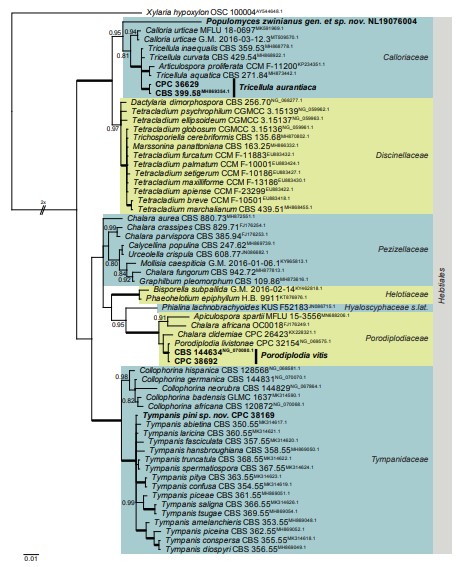
Fig. 2. Consensus phylogram (50 % majority rule) resulting from a Bayesian analysis of the Leotiomycetes LSU nucleotide alignment. Bayesian posterior probabilities (PP) > 0.79 are shown at the nodes and the scale bar represents the expected changes per site. Thickened branches represent PP = 1 and the most basal branch was halved to facilitate layout. Families and the order Helotiales are indicated with coloured blocks to the right of the tree. GenBank accession (superscript) and / or culture collection / voucher numbers are indicated for all species. The tree was rooted to Xylaria hypoxylon (voucher OSC 100004, GenBank AY544648.1) and the species treated in this study for which LSU sequence data were available are indicated in bold face.
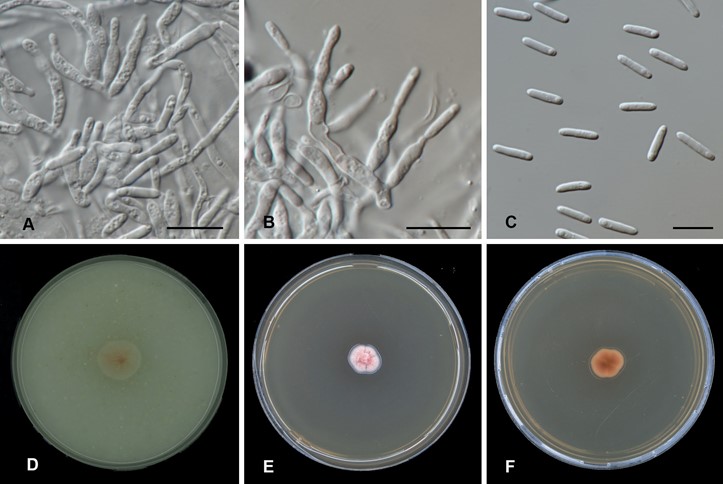
Fig. 59. Populomyces zwinianus (CBS 147307). A–B. Conidiophores and conidia. C. Conidia. D. Colony on OA. E. Colony on MEA. F. Reverse colony on MEA. Scale bars A–C = 10 μm.
