Oblongohyalospora macarangae Tennakoon, C.H. Kuo, S. Hongsanan & K.D. Hyde, sp. nov.
MycoBank number: MB 555437; Index Fungorum number: IF 555437; Facesoffungi number: FoF 09331, Fig. 41
Etymology: Name reflects the host Macaranga tanarius. Holotype: MFLU 18-0082
Saprobic on dead leaves petiole of Macaranga tanarius, appearing as small dark brown to black dots. Hyphae superficial, straight to substraight, dark brown, irregular, easily removed from the host, appressoria not observed. Sexual morph: Undetermined. Asexual morph: Pycnothyria 120–200 μm diam., superficial, scattered, rounded to oval, flattened, brown to black, opening by stellate fissures. Upper wall comprising an irregular arrangement of dark cells, cells at margin branching and forming superficial hyphae. Conidiogenous cells evanescent. Conidia 11–13.5 × 5–6.2 µm ( x̄= 12.5 × 5.5 μm, n = 30), unicellular, oblong, hyaline, dis- tinct 2–3 large guttules, straight or slightly curved, smooth walled.
Culture characteristics: Colonies on PDA, 20 mm diam. after 3 weeks, colonies from above: medium dense, raised, circular, convex, surface slightly smooth, entire edge, effuse, velvety to hairy, margin well-defined, dark brown to dark grey at the margin, grey to yellowish at the centre; reverse: dark brown to black at the margin, grey at the centre; mycelium light brown to olivaceous with tufting; yellowish pigment in PDA.
Material examined: Taiwan, Chiayi, Ali Shan Mountain, Fanlu Township area, Dahu forest, dead leaves petioles of Macaranga tanarius (Euphorbiaceae), 05 June 2019, D. S. Tennakoon, M024A (MFLU 18-0082, holotype); ex-type living cultures, MFLUCC 18-0552, NCYUCC 19-0226, ibid., 10 June 2019, M024B (NCYU 19-0356, paratype); ex-paratype living culture, NCYUCC 19-0329.
GenBank numbers: MFLUCC 18-0552: LSU: MW063241. NCYUCC 19-0329: LSU: MW063242.
Notes: – The asexual morphs of Asterinales are coelomycetous (pycnothyrial) or hyphomycetous and most of sexual-asexual connections have been established based on joint development on the same substrate (Hosagoudar 2012; Hyde et al. 2013; Hongsanan et al. 2014a). Oblongohyalosporaceae is described herein as a new family in Asterinales, to accommodate Oblongohyalospora. Oblongohyalospora species collected from dead leaves petioles of Macaranga tanarius and, are characterized by superficial, scattered, rounded to oval, flattened, brown to black pycnothyria, upper wall comprising an irregular arrangement of dark cells and distinct unicellular, oblong, hyaline conidia with 2–3 large guttules (Fig. 41). The upper wall of pycnothyria differs from the other families in Asterinales in having irregular arrangement of dark cells, whereas thyrothycia/ pycnothyria of most other families have linear, parallel arrangement of cells, which are radiating from the centre (Hongsanan et al. 2014). In our phylogeny, Oblongohyalospora species constitutes an independent lineage branching between Asterotexaceae and Neobuelliellaceae with 80% ML, 0.98 BYPP statistical support (Fig. 42). The sexual-asexual connection of Asterotexaceae and Neobuelliellaceae, is not established, since no asexual morphs from any species have been obtained (Hongsanan et al. 2020).
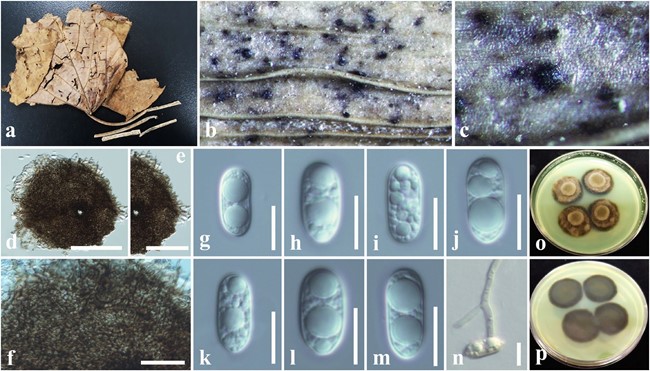
Fig. 41 Oblongohyalospora macarangae (MFLU 18-0082, holotype): a Dead leaf of Macaranga tanarius. b, c Appearance of colonies with pycnothyria on the leaf petiole. d Squash mount of pycnothyrium. e, f Upper walls. g–m Conidia. n A germinating conidium. o Colonies from above (on PDA). p Colonies from below (on PDA). Scale bars: d–f = 50 µm, g–n = 8 µm
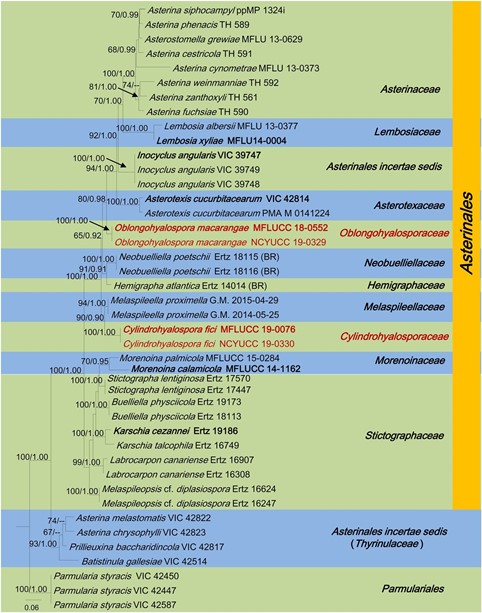
Fig. 42 The best scoring RAxML tree with a final likeli- hood value of – 6084.384150 for combined dataset of LSU sequence data. The tree is rooted with Parmularia styracis (VIC 42447, 42450, 42587). The matrix had 870 distinct alignment patterns with 4% undetermined characters and gaps. Estimated base frequen- cies were as follows; A = 0.239761, C = 0.245770, G= 0.323813, T = 0.190656;substitution rates AC = 0.926216, AG = 2.978501, AT= 0.762681, CG = 1.285420, CT = 9.230560, GT =1.000000; gamma distribution shape parameter α = 0.528622.Ex-type strains are indicated in bold. Newly generated sequences are in red. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes
