Neomonodictys Y.Z. Lu, C.G. Lin & K.D. Hyde
Index Fungorum number: IF556730; Facesoffungi number: FoF 06710
Etymology: Name reflects its morphological similarity to Monodictys.
Saprobic on decaying wood.
Sexual morph: Undetermined.
Asexual morph: Hyphomycetous, dictyosporous. Colonies on natural substratum superficial, scattered, black, glistening. Mycelium immersed in the substrate, composed of septate, branched, smooth, thin-walled, hyaline to pale brown hyphae. Conidiophores lacking. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, brown. Conidia acrogenous, solitary, subglobose to globose, muriform, pale brown when immature, darkened to black when mature, sometimes with one basal cell paler than the others.
Notes: – The monophyletic asexual genus Neomonodictys is established for a fungus collected from a freshwater habitat in Thailand that is morphologically similar to members of Monodictys S. Hughes, but phylogenetically distinct (Fig. 132). Monodictys is characterised by monoblastic, hyaline to brown conidiogenous cells, solitary, dictyospores, subglobose, pyriform or clavate conidia (Ellis 1971; Seif- ert et al. 2011). Conidiophores of Monodictys are often reduced to conidiogenous cells (Ellis 1971; Seifert et al. 2011). Monodictys were placed in Dothideomycetes (Day et al. 2006; Seifert et al. 2011; Wijayawardene et al. 2017a, 2018a). Phylogenetic inference in this study showed that Neomonodictys muriformis formed a highly supported clade within Pleurotheciaceae (Pleurotheciales, Sordariomycetes) (Fig. 132), which is distant to Monodictys. Therefore, the new generic name, Neomonodictys, is introduced.
Type species: Neomonodictys muriformis Y.Z. Lu, C.G. Lin & K.D. Hyde.
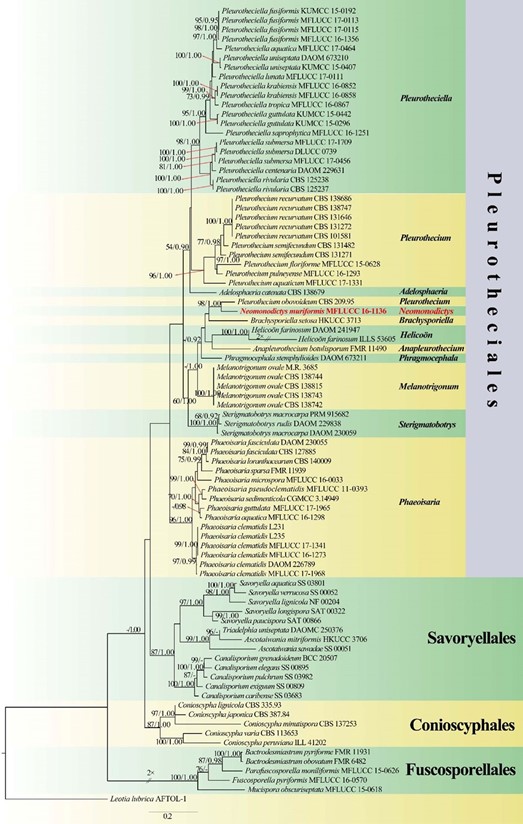
Fig. 132 Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data representing Pleu- rotheciales and the closely related orders. Eighty-five strains are included in the combined analyses which comprise 1871 characters (1194 characters for LSU, 677 characters for ITS) after alignment. Leotia tubrica (AFTOL-1) is used as the outgroup taxon. The best RAxML tree with a final likelihood values of − 18609.630764 is pre- sented. The matrix had 1200 distinct alignment patterns, with 30.71% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.231344, C = 0.256609, G = 0.300675, T = 0.211372; substitution rates AC = 1.000000, AG = 3.698119, AT = 1.000000, CG = 1.000000, CT = 3.698119, GT = 1.000000; gamma distribution shape parameter α = 0.309377. Bootstrap values for maximum likeli- hood (ML) equal to or greater than 75% and clade credibility values greater than 0.90 from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indi- cated in bold and red
Species
