Montanitestudina Maharachch., Wanas. & Al‑Sadi, gen. nov.
MycoBank number: MB 837558; Index Fungorum number: IF 837558; Facesoffungi number: FoF 09487;
Etymology: The generic epithet is from the combination of two words “montānus” (mountain) habitat where the fungus was isolated and “testudina” similar to Testudina. Saprobic on dead wood of unknown plants.
Sexual morph: Ascomata scattered or sometimes gregarious beneath the host periderm or on decorticated wood, coriaceous, black, globose to subglobose, ostiolate. Ostiole central, papillate, with an irregular, pore-like opening. Peridium composed of 4–5-layers with brown to dark-brown, cells of textura angularis fusing the host tissues. Hamathecium comprising septate, cellular pseudoparaphyses. Asci 8-spored, bitunicate, fissitunicate, cylindrical to cylindric-clavate, with a distinct pedicel, apically rounded, with an ocular chamber. Ascospores overlapping uniseriate, brown, ellipsoid, oblong to fusoid, 3–5-transversely septate, with 1–2-longitudinal septa, muriform, smooth-walled, with or without surrounded by a mucilaginous sheath. Asexual morph: Not observed.
Type: Montanitestudina hydei Maharachch., Wanas. & Al-Sadi
Notes:—Montanitestudina morphologically resembles some of the species in Camarosporidiella Wanas. et al., Cucurbitaria Gray, Fenestella Tul. & C. Tul., Hawksworthiana U. Braun, Marjia Wanas. et al., Neocucurbitaria Wanas. et al., Pseudostrickeria Q. Tian et al., Sporormurispora Wanas. et al., and Uzbekistanica Wanas. et al. by its cylindrical asci and uniseriate, brown, muriform ascospores (Wanasinghe et al. 2017, 2018; Jaklitsch et al. 2018). Although there is some morphological overlap between Montanitestudina and the above-mentioned genera, they are phylogenetically apart from Montanitestudina in multi-gene phylogenetic analyses. Montanitestudina is phylogenetically closely related to Lepidosphaeria (Fig. 8). However, this is strongly supported only by Bayesian analysis. There are substantial morphological differences between these two taxa to warrant generic ranks.
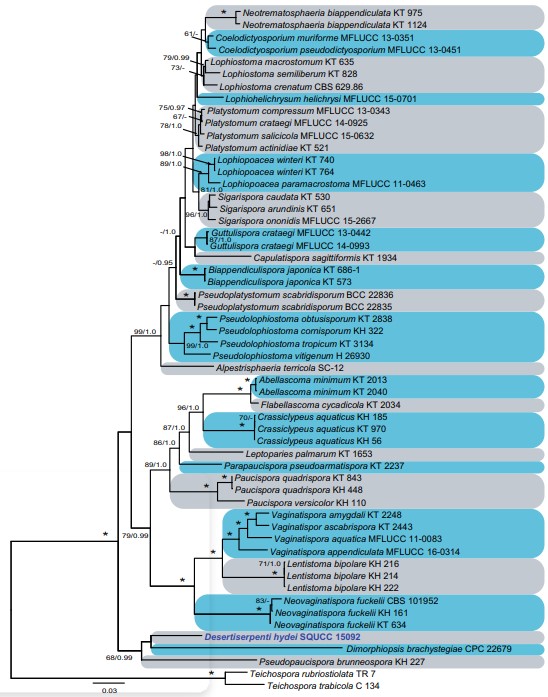
Fig. 8 Phylogenetic tree inferred using the combined sequences of LSU, SSU, ITS, RPB2 and TEF of the analysed Lophiostomataceae genera. The tree backbone was constructed using maximum likelihood (ML) analysis. The MLB and PP above 60% and 0.95, respectively, are given at the nodes (MLB/PP). An asterisk (*) indicates branches with MLB = 100% and PP values = 1.0. Species are followed by the strain accession numbers. The new strain is indicated in blue bold. The scale bar represents the expected number of changes per site for ML
Species
