Melanconis stilbostoma (Fr.) Tul. & C. Tul., Selecta fungorum carpologia (Paris) 2, 115 (1863)
Index Fungorum number: IF 243057; Facesoffungi number: FoF 04448; Fig. 1
Plant pathogenic on the twigs of Betula pubescens. Asexual morph: Conidiomata 1100–1300 × 550–650 µm (x̄ = 1257 × 592 µm, n = 10), stromatic, acervular or pulvinate, solitary to aggregated, immersed to semi-immersed, unilocular with circular covering disc, slightly convex, yellowish-white. Conidiomatal wall 80–200 µm wide, surrounded and intermingled with host cells, thick-walled. Hymenium comprises dense, hyaline hyphae with the cells of textura intricata. Conidiophores start from a pseudoparenchymatous base, filiform, aseptate, branched at the base, smooth-walled, strongly aggregated, hyaline at the beginning, and turn brown from the tips. Conidiogenous cells highly dense, originating from the terminal of conidiophores, annellidic, enteroblastic, cylindrical, repetitive, producing hyaline α-conidia. Conidia 15–25 × 8–15 µm (x̄ = 20 × 11 µm, n = 20, l/w = 1.8), oval, ellipsoidal to subglobose, aseptate, thick-walled, smooth, hyaline at immaturity, pale brown to brown at maturity without β-conidia. Sexual morph: see Jaklitsch & Voglmayr (2020).
Material examined – Russia, Rostov region, Shakhty City District, Lenin Street, urban trees, on dying (dieback) twigs of Betula pubescens, 8 May 2018, Timur S. Bulgakov, T-7510 (MFLU 18-1885).
Known distribution (based on molecular data) – Austria, China, Italy, Japan, Poland, Russia, Sweden, United States and Ukraine (Zhuang 2005, Sogonov et al. 2008, Voglmayr et al. 2012, Fan et al. 2016, Hyde et al. 2020c, Jaklitsch & Voglmayr 2020, Farr & Rossman 2023, this study).
Known hosts (based on molecular data) – Betula spp. (B. aetnensis, B. pendula, B. platyphylla var. japonica, B. platyphylla, B. papyrifera, B. rotundifolia and B. tianschanica) (Zhuang 2005, Sogonov et al. 2008, Voglmayr et al. 2012, Fan et al. 2016, Hyde et al. 2020c, Jaklitsch & Voglmayr 2020, Farr & Rossman 2023), Betula pubescens (this study).
GenBank accession numbers – ITS: OQ629853, LSU: OQ629854, rpb2: OQ653555, tef1-α: OQ653556
Notes – In the multi-gene phylogeny, our strain MFLU 18-1885 clustered with strains of Melanconis stilbostoma (CBS 109778, D 143, MAFF 410225, CFCC 50477, CFCC 50476, CFCC 50475, CFCC 50478 and D 258) with 100 % maximum likelihood bootstrap support and 1.00 Bayesian posterior probability (Fig. 2). Our collection shares similar morphologies to the asexual morph of M. stilbostoma described by Jaklitsch & Voglmayr (2020). Melanconis stilbostoma is widespread in European and Asian parts of Russia and adjacent countries in natural habitats of Betula species (Sinadsky 1973, Cherepanova & Cherepanov 2003, Rebriev et al. 2012). Previously, M. stilbostoma was recorded and confirmed on B. pendula in Donetsk (Hyde et al. 2020c) and Lugansk regions (Ordynets et al. 2013). Based on morphology (Fig. 1) and molecular data analysis (Fig. 53), we conclude our collection is the first record of M. stilbostoma on Betula pubescens in Rostov region, Russia.
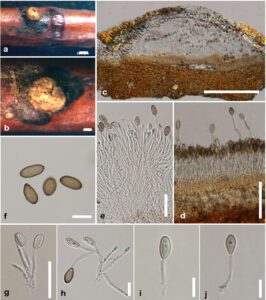
Fig. 1– Melanconis stilbostoma (MFLU 18-1885, a new host record). a–b A conidioma on twigs of Betula pendula. c Longitudinal section of a conidioma. d–e Arrangement and development of conidiospores and conidiogenous cells inside conidiomata. g–j Conidia. Scale bars: a, c = 500 μm, b = 200 μm, d = 100 μm, e, g = 50 μm, f, h–j = 20 μm.
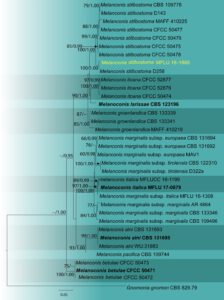
Fig. 2 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, rpb2 and tef1-α sequence data. Thirty-five strains were included in the combined sequence analyses, which comprised 3268 characters with gaps (ITS = 528, LSU = 858, rpb2 = 1161, tef1-α = 721). Single gene analyses were also performed, and topology and clade stability compared with the combined gene analyses. Gnomonia gnomon (CBS 829.79) strain was used as the outgroup taxon. Final ML optimization likelihood is -7101.950102. The matrix included 311 distinct alignment patterns, with 11.83 % undetermined characters or gaps. Estimated base frequencies were obtained as follows: A = 0.247189, C = 0.263714, G = 0.268910, T = 0.220187; substitution rates AC = 1.490211, AG = 3.125914, AT = 1.908995, CG = 0.756262, CT = 9.270114, GT = 1.0; gamma distribution. Bootstrap support values for ML (first set) equal to or greater than 65%, BYPP equal to or greater than 0.95 are given above or below the nodes. The strain from the current study is in yellow and the type strains are in bold.
