Marasmius obscuroaurantiacus Wannathes, N. Suwannarach, J. Kumla & Lumyong, sp. nov.
MycoBank number: MB 839832; Index Fungorum number: IF 839832; Facesoffungi number: FoF 10617; Fig. 22
Etymology: ‘obscuro’ = dusky and ‘aurantiacus’ = orange color, refers to the dusky orange coloured pilei.
Holotype: BKF10242
Pileus 2–5 mm diam., convex with tiny dark brown conical papilla at center when young, expanding to plano-convex, umbilicate, with a rounder papilla with age; dull, dry, glabrous, striate; orange (5B8) at disc greyish orange (5B5) at margin when young, brownish orange (6C6–7) at disc, greyish orange (5B5) at margin with age. Context thin, pale orange (5A3). Lamellae adnate to a small collarium, distant (7–9), narrow (0.5 mm), orange white (5A2). Stipe 18–26 × 0.3 mm, cylindrical, central, glabrous, dull, dry, insititious, brown overall, some stipe arising directly from dark brown rhizomorphs. Basidiospores 7–8(–9) × 3–4 mm [x̅ = 7.36 ± 0.57 × 3.52 ± 0.39, Q = 1.75–2.57, q = 2.11 ± 0.26, n = 25], ellipsoid, pyriform, smooth, hyaline, inamyloid, thin-walled. Basidia 17–20 × 6–7 mm, clavate, with 4 sterigmata, thin-walled, inamyloid. Cheilocystidia of Siccus-type broom cells; main body 8–12 × 6–8 µm, clavate to pyriform, hyaline, inamyloid, thin-walled; apical setulae 1–4 × 1 µm, cylindrical to conical, obtuse, hyaline, thick-walled. Pleurocystidia absent. Pileipellis hymeniform, mottled, composed of Siccus-type broom cells; main body 7–13 × 5–10 µm, clavate to pyriform or turbinate, hyaline, inamyloid, thin-walled; apical setulae 1–4 × 1 µm, cylindrical to conical, obtuse, brown, thick-walled. Pileus trama interwoven, inamyloid. Lamellar trama hyphae 3–10 µm diam., interwoven, cylindrical, smooth, hyaline, inamyloid, thin-walled, non-gelatinous. Stipitipellis hyphae 3–5 µm diam., subparallel, cylindrical, brown, smooth, inamyloid, thick-walled (up to 1–1.5 µm), non-gelatinous. Stipe trama hyphae 3–4 µm diam., subparallel, cylindrical, hyaline, smooth, inamyloid to weak dextrinoid, thin-walled, non-gelatinous. Caulocystidia absent. Clamp connections present in almost all tissues, absent at the hyphae of stipitipellis.
Habitat: gregarious on dicotyledonous leaves.
Distribution: Thailand.
Material examined: Thailand, Nakhon Ratchasima Province, Khao Yai National Park, trail to Pha Kluai Mai waterfall, 21 Sep 2018, N Wannathes, N Suwannarach and J Kumla, (BKF10242, holotype).
GenBank numbers: ITS: MZ145165.
Notes: Marasmius obscuroaurentiacus is morphologically similar to M. crinisequi and M. trichorhizus. Marasmius crinisequi differs by forming shorter stipe and longer basidiospores (Shay et al. 2017). Marasmius trichorhizus differs by forming pileipellis composed of mixed Siccus-type broom cells and non-setulose cells (Oliveira et al. 2020). Marasmius obscuroaurentiacus differs by 227 and 236 base pairs in the ITS region compared to M. crinisequi and M. trichorhizus, respectively. Phylogenetic analyses inferred from ITS sequence data confirm that M. obscuroaurentiacus is a distinct species (Fig. 23).
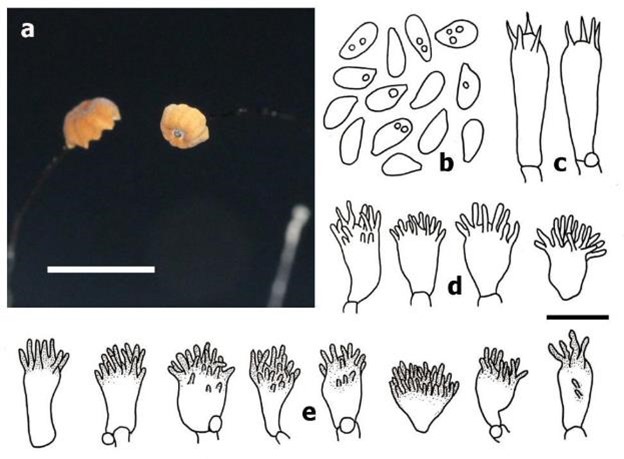
Fig. 22 Marasmius obscuroaurantiacus (BKF10242, holotype). a Basidiomata. b Basidiospores. c Basidium. d Cheilocystidia. e Pileipellis cell. Scale bars: a = 10 mm, b–e = 10 μm.
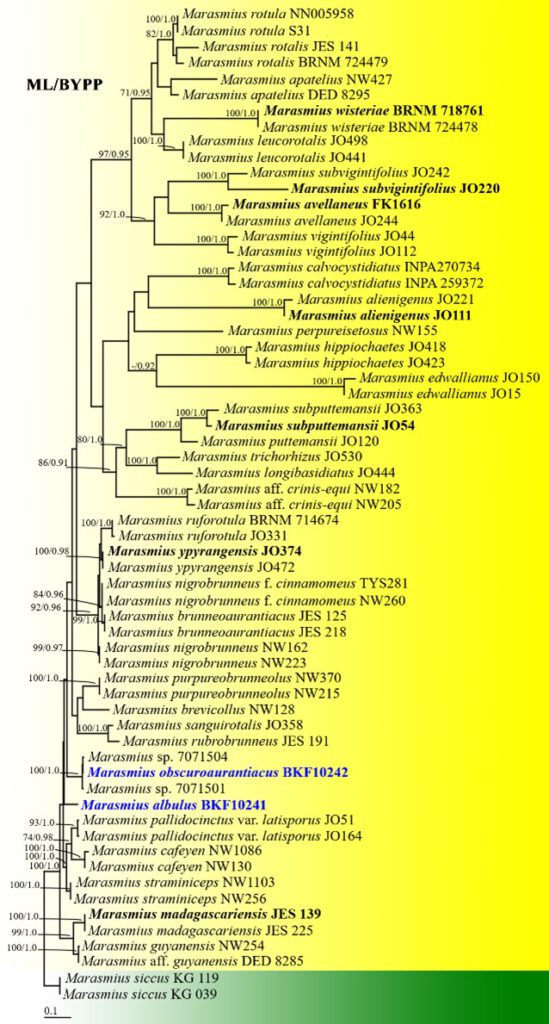
Fig. 23 Phylogenetic tree derived from ML analysis of ITS region of 63 sequence data and the aligned dataset comprised 1217 characters including gap. The average standard deviation of the split frequencies of the BYPP analysis was 0.00621. The best scoring RAxML tree had a likelihood value of -12303.3407. The matrix had 849 distinct alignment patterns with 45.66% undetermined characters or gaps. Estimated base frequencies were: A = 0.2423, C = 0.2022, G = 0.2224, T = 0.3310; substitution rates AC = 0.9731, AG = 3.0198, AT = 1.0578, CG = 0.85296, CT = 3.3441, GT = 1.0000. Marasmius siccus strains (KG 039 and KG 119) were used as outgroups. Maximum likelihood bootstrap values ≥ 70% and BYPP greater than 0.90 are given at the nodes. The bar represents 0.1 substitutions per nucleotide position. Hyphen (-) represents support values ≤70%/0.90 BYPP. The ex-type strains are in bold and black. The newly generated sequences are indicated in bold and blue.
