Lepiota angusticystidiata J.F. Liang & Z.L. Yang, in Liang, Yu, Lu, Wang & Song, Mycologia 110, 496 (2018)
Index Fungorum number: IF 819510; Facesoffungi number: FoF 06198; Figs. 1, 2
Pileus 10–45 mm diam., first irregular subglobose to conical, expanding to parabolic to campanulate, umbonate, becoming plano-convex to concave when fully mature, with straight or slightly inflexed margin; covered with light brown to dark brown (6D7-6, 6E6–7, 6F6-8) granulose when young, surface breaking up and living concolorous granulose umbo, with light brown to brown (6D7-6, 6E6–7) squamules around umbo towards the margin on white to yellowish white (4A2) background; margin cortinate, with concolorous squamules, sometimes fringed. Lamellae free, white, crowded, ventricose, 2–5 mm wide, with concolorous eroded edge. Stipe 20–80 × 4–8 mm, cylindrical, narrow at apex, slightly wider at base; with concolorous squamules as those on pileus from annular zone towards base on with white to yellowish white (4A2) background. Annulus an annular zone, not well-defined, with white fibrillose and concolorous squamules. Context white in pileus, 2–4 mm wide; concolorous with surface in stipe, hollow. Smell and Taste not observed. Spore print white.
Basidiospores [50,2,2] 6.3–7.2 × 4.0–5.3 μm, avl × avw = 6.81 × 4.50 μm, Q = 1.36–1.63, avQ = 1.51, ellipsoid ovoid in sideview, ellipsoid in frontal view, thick-walled, hyaline, dextrinoid, congophilous, cyanophilous, not metachromatic. Basidia 19–23 × 8–10 μm, clavate, thick-walled, hyaline, 4-spored, rarely 2-spored. Pleurocystidia absent. Cheilocystidia 10–30 × 5–11 μm, abundant, mostly utriform to narrowly utriform or fusiform, sometimes clavate, branched and septate, hyaline, thick-walled. Pileus covering a trichoderm made up of cylindrical elements with attenuated apex, 100–270 × 7–18 μm, septate and branch at base, thick-walled, intracellular and with parietal pale brown pigment, with concolorous narrowly clavate elements under long elements, 35–60 × 10–23 μm. Stipe covering a trichoderm similar to pileus covering. Clamp-connections present in all tissues.
Material examined – Laos, Vientiane Capital, Xaythany District, Houynhang Preserve Forest, 9 July 2016, P. Sysouphanthong (HNL503160); ibidem., 7 July 2016, P. Sysouphanthong (HNL503157); ibidem., 04 Aug. 2016, P. Sysouphanthong (HNL503163).
GenBank accession numbers – HNL503160 – ITS: MZ717725.
Known habitat – grow solitary to a large group, terrestrial on soil mixed with decayed leaves and branches (Liang et al. 2018, Hyde et al. 2020b).
Known distribution (based on molecular data) – China (Liang et al. 2018), northern Thailand (Hyde et al. 2020b), central Laos (this study).
Note – According to the morphological characteristics, Lao specimens of Lepiota angusticystidiata are similar to type specimens from China (Liang et al. 2018). Thai specimens have longer pileus and stipe covering, but other characters are similar to Lao and China specimens (Hyde et al. 2020b). The phylogenetic analysis of nrITS sequences indicated that specimens of China, Laos and Thailand are clustered with 98% bootstrap support (Fig. 3). Our species is related to the clade of Lepiota sect. Ovissporae, which consists of species with long elements, and species with short clavate elements in Lepiota sect. Lepiota (Figs. 1 & 2). Some species related to L. angusticystidiata were discussed in Liang et al. (2018) and Hyde et al. (2020b). Current study presents a new geographical record of Lepiota angusticystidiata from central Laos.

Fig. 1 – Basidiomata of Lepiota angusticystidiata in situ (HNL503160, new country record). a–b HNL503163. c HNL503160. d–e HNL503157.
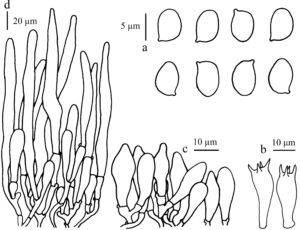
Fig. 2 – Micro-characteristics of Lepiota angusticystidiata (HNL503160). a Basidiospores. b Basidia. c Cheilocystidia. d. Elements structure on pileus covering.
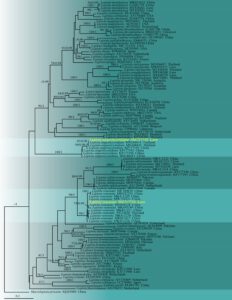
Fig. 3 – Maximum likelihood phylogenetic tree (ML) of Lepiota based on nrITS sequences. One hundred and eight strains consisting of 683 characters after alignment (including gaps). The tree topology derived from the Bayesian analysis was similar to that derived from the ML analysis. The best RaxML tree with a final likelihood value of -11952.807294 is presented. The matrix had 474 distinct alignment patterns, with 7.95% undetermined characters or gaps. Evolutionary model applied is GTRGAMMA. Bootstrap values of ML equal to greater than 70% and Baysian posterior probabilities equal to greater than 0.94 are given above branches. The tree is rooted with Macrolepiota procera (JZB2115036). Newly generated sequence is indicated in yellow.
