Hilberina Huhndorf & A.N. Mill., Mycol. Res. 108(1): 31 (2004)
Index Fungorum number: IF 28830; MycoBank number: MB 28830; Facesoffungi number: FoF 13209; 17 morphological species (Species Fungorum 2020), 5 species with sequence data.
Type species – Hilberina caudata (Fuckel) Huhndorf & A.N. Mill.
Notes – Hilberina caudata has setose ascomata, cylindrical asci with rings and geniculate ascospores with one end tapering to a distinct point (Miller & Huhndorf 2004). Sequence data of five members in Hilberina were provided by Miller et al. (2014) and revealed that Hilberina was closely related to Synaptospora, Ruzenia and Helminthosphaeria within Helminthosphaeriaceae (Fig. 8).

Figure 8 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Chaetosphaeriales and Tracyllalales taxa. Ninety-six strains are included in the combined analyses which comprised 1695 characters (1081 characters for LSU, 614 characters
for ITS) after alignment. Neurospora crassa MUCL 19026 and Gelasinospora tetrasperma CBS 178.33 (Sordariaceae, Sordariales) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 23777.689886 is presented. Estimated base frequencies were as follows: A = 0.231060, C = 0.264793, G = 0.308265, T = 0.195882; substitution rates AC = 1.388486, AG = 1.836207, AT = 1.649563, CG = 0.971659, CT = 6.316962, GT = 1.000000; gamma distribution shape parameter a = 0.460297. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
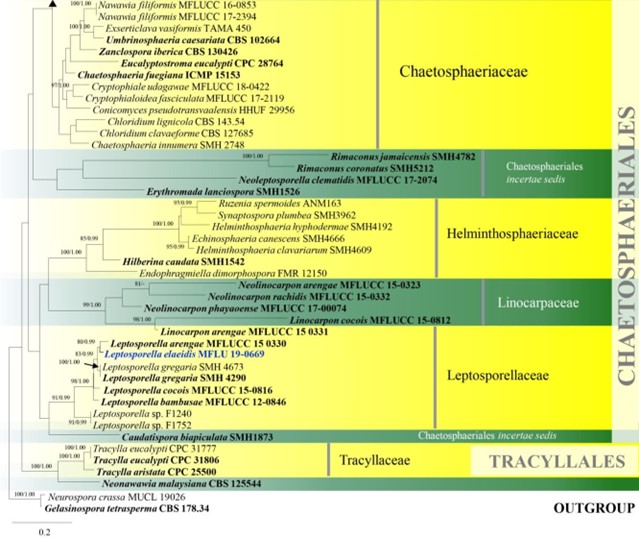
Figure 8 – Continued.
Species
