Cryphognomonia pini C.M. Tian & N. Jiang
MycoBank number: MB 829510; Index Fungorum number: IF 829510; Facesoffungi number: FoF14425; Figure 4
Diagnosis. Cryphognomonia pini differs from its closest phylogenetic neighbor, F. rhoigena, in ITS, LSU, tef1 and rpb2 loci based on the alignments deposited in TreeBASE.
Etymology. Named after the genus of the host plant from which the holotype was collected, Pinus.
Description. Pseudostromata erumpent, causing a pustulate bark surface, 650– 1200 µm diam., containing up to 12 perithecia. Central column yellowish to brownish. Stromatic zones lacking. Perithecia conspicuous, flask-shaped to spherical, umber to fuscous black, regularly scattered, 350–600 µm diam. Paraphyses deliquescent. Asci fusoid, 8-spored, biseriate, with an apical ring, (60–)65–80(–90) × (21–)22– 31(–35) µm. Ascospores hyaline, clavate to cylindrical, smooth, multi-guttulate, symmetrical to asymmetrical, straight to slightly curved, bicellular, with a median septum distinctly constricted, with distinct hyaline sheath, (15.5–)18–25(–27) × (8.5–)9.5– 11.5(–12) µm. Asexual morph: not observed.
Culture characters. Cultures incubated on PDA at 25 °C in the dark, initially pale white, becoming olive-green after 3 wk. The colonies are flat, with regular margins; texture initially uniform, becoming compact after 1 month.
Specimens examined. China. Shaanxi Province: Ankang City, Huoditang forest farm, 33°26’7″N, 108°26’48″E, on branches of Pinus armandii, 8 June 2018, N. Jiang & C.M. Tian (holotype BJFC-S1725; ex-type living culture: CFCC 53020); 33°26’7″N, 108°26’48″E, on branches of Pinus armandii, 8 June 2018, N. Jiang & C.M. Tian (BJFC-S1726; living culture: CFCC 53021).
Notes. Cryphognomonia pini is the type species of Cryphognomonia, and occurs on Pinus armandii in China. Morphologically, Cryphognomonia pini is characterized based on bicellular ascospores with obvious hyaline sheath. In the phylogenetic tree, this species is most closely related to F. rhoigena (Fig. 1). However, Cryphognomonia pini can be distinguished from F. rhoigena based on ITS, LSU, tef1 and rpb2 loci (73/512 in ITS, 4/775 in LSU, 186/437 in tef1 and 90/1064 in rpb2).
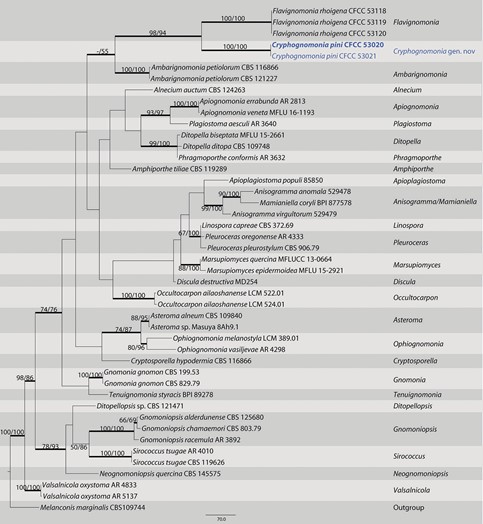
Figure 1. Maximum parsimony phylogram of Gnomoniaceae based on a combined matrix of ITS, LSU, tef1 and rpb2 genes. The MP and ML bootstrap support values above 50% are shown at the first and second position, respectively. Thickened branches represent posterior probabilities above 0.90 from BI. Scale bar: 80 nucleotide substitutions. Strains in this study are in blue and ex-type strains are in blod.
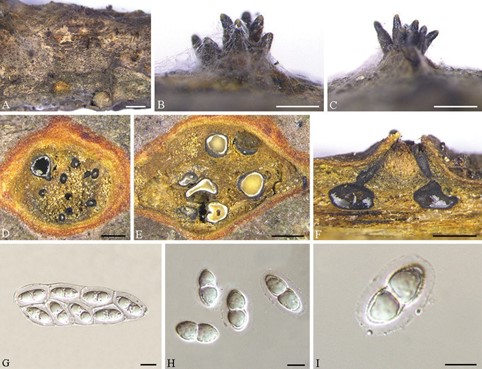
Figure 4. Cryphognomonia pini on Pinus armandii (BJFC-S1725) A–C habit of ascomata on twigs D, E transverse section of ascomata F longitudinal section through ascomata G asci H, I ascospores. Scale bars: 2 mm (A); 500 µm (B–F); 10 µm (G–I).
