Colletotrichum orchidis Jayaward., Camporesi & K.D. Hyde, sp. nov. Fig. 108
MycoBank number: MB 556214; Index Fungorum number: IF 556214; Facesoffungi number: FoF 05784;
Etymology – Refers to the host genus.
Holotype – MFLU 16-2551.
Saprobic on aerial stem of Orchis sp. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 55–135 μm ( x = 95 μm, n = 10) diam., black, acervulus, oval, solitary or gregarious, comprising dark brown, roundish cells from which setae and conidiophores develop. Setae straight or ± bent, abundant, dark brown to light brown, becoming paler towards the apex, smooth-walled, 3–5-septate, 105 μm long, base cylindrical, slightly inflated, 5. 6 μm diam, apex acute to rounded. Chlamydospores not observed. Conidiophores simple, to 65 μm long, hyaline to pale brown, smooth-walled. Conidiogenous cells 8–11 × 3.1–4.4 μm ( x = 10.5 ± 1.8 × 3.6 ± 0.4 μm, n = 20), hyaline, smooth-walled, cylindrical to slighty inflated, opening 0.5–1 μm wide, collarette or periclinal thickening not observed. Conidia 15.9–20.3 × 1.9–3.2 μm ( x = 18.2 ± 1.5 × 2.5 ± 0.6 μm, n = 40), L/W ratio 7.3, hyaline, smooth or verruculose, aseptate, very variable in size and shape, some strongly curved, strongly curved towards the often broadly rounded apex than towards the truncate base, some small conidia almost straight, guttulate. Appressoria 8.2–13.1 × 4.5–8.2 μm ( x = 11.5 ± 3.2 × 4.9 ± 1.4 μm, n = 10), L/W ratio 2.3, solitary or in loose groups, single-celled, olivaceous brown to dark brown, irregularly-shaped, but often globose or clavate, smooth-walled.
Culture characteristics – Colonies on PDA flat with entire margin, aerial mycelium sparse, short, pale olivaceous-grey, iron-grey acervuli can be observed mainly on the edge of the colony after 7 days; reverse olivaceous green, reaching 60–75 mm in 7 d at 18 °C.
Material examined – ITALY, Province of Forlì-Cesena, near Premilcuore, on living dead aerial stem of Orchis sp. (Orchidaceae), 5 October 2016, E. Camporesi IT3118 (MFLU 16-2551, holotype), extype-living culture MFLUCC 17-1302; ibid cultures KUMCC 17-0119, JZB330118.
GenBank numbers – ITS: MK502144, gapdh: MK496857, CHS: MK496855, act: MK496853, tub2: MK496859 (MFLUCC 17-1302), ITS: MK502143, gapdh: MK496858, CHS: MK496856, act: MK496854, tub2: MK496860 (JZB330118).
Notes – Colletotrichum orchidis is only known from aerial stem of Orchis in Italy. The conidia of this species resemble those of several species complexes in Colletotrichum (e.g. dematium species complex, graminicola species complex, spaethianum species complex and truncatum species complex) (Jayawardena et al. 2016). Based on DNA sequences, C. orchidis falls within the C. dematium species complex clade and forms a separate branch as a sister taxon of C. dematium ( Fig. 107) . A BLASTn search of NCBI GenBank with the ITS sequence of the new species, showed 96% similarity to several Colletotrichum species with curved conidia. The closest match in a BLASTn search in GenBank with the gapdh sequence of the new species showed 99% similarity to C. dematium (2bp differences) and C. lineola (4bp differences). CHS sequence showed a 98% similarity to C. dematium ( 6 bp differences) and C. insertae ( 7 bp differences) and act sequences also showed a 98% similarity to C. dematium (4 bp differences). tub2 sequence showed 99% similarity to C. dematium (1bp differences) and C. lineola (1 bp difference). Colletotrichum orchidis differs from C. dematium by smaller conidia and a lower L/W value (C. dematium 21.3 × 3.5, L/W = 6.0).

Figure 108 – Colletotrichum orchidis (MFLUCC 17-1302, holotype). a-c Appearance of conidiomata on host. d Setae. e Base of seta. f, h Conidiophores. g Conidiogenous cell. i-k Conidia. l-n Appressoria. o 3 day old culture on PDA. p 10 day old culture on PDA. Scale bars: b, c = 200 µm, d, f, h = 20 µm, e, g, i-k, l-n = 10 µm.
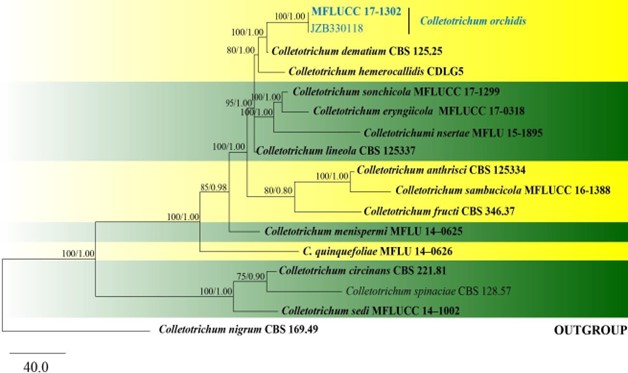
Figure 107 – One of the 100 most phylogenetic tree generated by maximum parsimony analysis of combined ITS, GAPDH, CHS, ACT and β-tubulin sequenced data of dematium species complex. Seventeen strains are included in the analyses, which comprise 1780 characters including gaps (509 characters for ITS, 267 characters for GAPDH, 250 characters for CHS, 237 characters for ACT, 501 characters for β-tubulin) after alignment. Colletotrichum nigrum (CBS 169.49, Glomerellaceae, Glomerellales) is used as the outgroup taxon and the tree is rooted with. Single gene analyses were carried out and the topology of each tree had clade stability. Tree topology of the maximum parsinmony analysis is similar to Bayesian analysis. The maximum parsimonious dataset consisted of 1182 constant, 343 parsimony-informative and 255 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum often equally most parsimonious trees with a length of 939 steps (CI=0.804, RI=0.774, RC=0.623, HI=0.196) in the first tree. Bootstrap support values for MP greater than 75% and Bayesian posterior probabilities greater than 0.90 are given near nodes respectively. Ex-type strains are in bold and black. The newly generated sequences are indicated in blue.
