Codinaea Maire, Publ. Inst. Bot. 3(4): 15 (1937)
MycoBank number: MB 7720; Index Fungorum number: IF 7720; Facesoffungi number: FoF 13053; 16 morphological species (Species Fungorum 2020), 4 species with sequence data.
Type species – Codinaea aristata Maire
Notes – Codinaea was introduced based on C. aristata (Maire 1937). Gamundi et al. (1977) examined several specimens of Dictyochaeta fuegiana and proposed Codinaea as a synonym of Dictyochaeta. Their treatment was accepted by most authors and many Codinaea taxa have been moved to Dictyochaeta (Kirschner & Chen 2002, Cruz et al. 2008, Liu et al. 2016, Whitton et al. 2012). However, re-examination of the two collections, on which Gamundí et al. (1977) based their description of D. fuegiana (LPS 38629, LPS 38630), revealed a fungus was misidentified as the type of Dictyochaeta fuegiana (Réblová 2004). Réblová (2004) re-examined the type specimen of Dictyochaeta fuegiana and gave detailed descriptions. Molecular studies by Réblová & Winka (2000) revealed that Dictyochaeta species with or without setulae clustered into distinct subgroups in the phylogenetic tree. Réblová (2000) proposed to retain Codinaea for species with setulae and placed species without setulae in Dictyochaeta. Seifert et al. (2011) and Li et al. (2012) accepted Réblová (2000) suggestion on segregation of Codinaea and Dictyochaeta as separate genera delineated by the presence or absence of conidial setulae and accepted both Codinaea and Dictyochaeta as valid genera.
Wijayawardene et al. (2018a) and Hernández-Restrepo (2017) also treated them as two distinct genera. Currently, 104 species of Dictyochaeta and 53 species of Codinaea have been described (Index Fungorum 2020). However, only few have DNA sequence data. There are no ex- type sequence data are available for the generic types of Codinaea or Dictyochaeta. Our phylogenetic analysis based on the available sequences of Dictyochaeta and Codinaea species, revealed that species of both genera are polyphyletic within Chaetosphaeriaceae (Fig. 8). Therefore, a detailed morpho-molecular analysis with more taxonomic sampling would be required to finally confirm the phylogenetic status of Codinaea and Dictyochaeta. Here we keep the Codinaea and Dictyochaeta as separate genera until the generic types of Codinaea and Dictyochaeta are recollected, sequenced and confirmed the link by molecular data.

Figure 8 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Chaetosphaeriales and Tracyllalales taxa. Ninety-six strains are included in the combined analyses which comprised 1695 characters (1081 characters for LSU, 614 characters for ITS) after alignment. Neurospora crassa MUCL 19026 and Gelasinospora tetrasperma CBS 178.33 (Sordariaceae, Sordariales) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 23777.689886 is presented. Estimated base frequencies were as follows: A = 0.231060, C = 0.264793, G = 0.308265, T = 0.195882; substitution rates AC = 1.388486, AG = 1.836207, AT = 1.649563, CG = 0.971659, CT = 6.316962, GT = 1.000000; gamma distribution shape parameter a = 0.460297. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
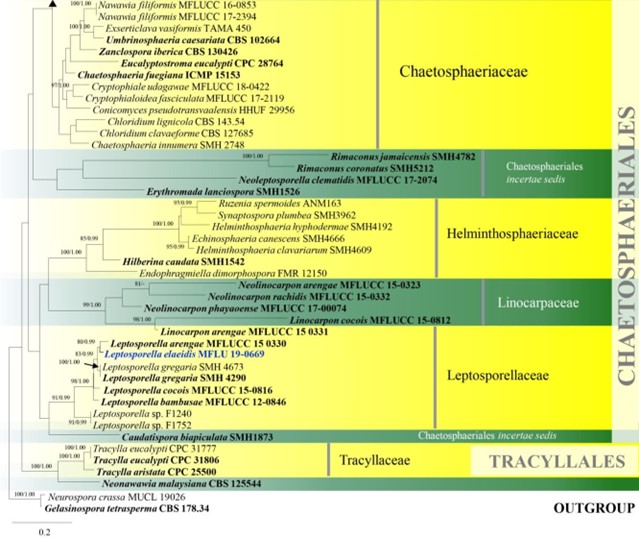
Figure 8 – Continued.
Species
