Arthrinium neogarethjonesii D.Q. Dai & K.D. Hyde, in Hyde et al., Mycosphere 11(1): 424 (2020)
MycoBank number: MB 555481; Index Fungorum number: IF 555481; Facesoffungi number: FoF 05112;
Etymology – Based on the similarities with species Arthrinium garethjonesii.
Holotype – HKAS 102408.
Saprobic on dead bamboo culms. Sexual morph: Stromata 1–2 mm long, 0.25–0.5 mm wide, 175–250 μm high, scattered to gregarious, immersed, becoming slightly erumpent through the host tissue, black to dark brown, fusiform to elongated ellipsoid, with a central slit-like opening at the top. Ascomata perithecial, 125–230 μm high, 120–230 μm diam., arranged in rows, clustered, gregarious, with 2–7 perithecia forming groups immersed in stromata and surrounding by stromatic tissue, ellipsoidal to subglobose, or obpyriform with a slightly flattened base, brown to reddish- brown, membranous. Ostiole raised from the centre of perithecia, internally lined with periphyses. Peridium 15–25 μm thick, composed of dark brown to reddish brown to hyaline cells of textura angularis. Stromatic cells between perithecia 150–350 μm wide, comprising host and fungal tissues, composed of elongated, brown cells of textura angularis. Paraphyses dense, long, 3.5–6 μm wide, septate, un-branched with a wider base and thinner apex. Asci 95–125 × 20–25 μm (x̅ = 97.6 × 21.3 μm, n = 20), 8-spored, unitunicate, clavate, apedicellate, apically rounded. Ascospores 25–30 × 9.5–11 μm (x̅ = 29.1 × 10.3 μm, n = 20), 2-seriate, overlapping, 1-septate, ellipsoidal, with a small lower cell and a large upper cell, with many guttules, hyaline, smooth-walled, with a thick gelatinous sheath, 3–10 µm wide. Asexual morph: Coelomycetous or Hyphomycetous on substrates, Conidiomata 450–600 µm wide × 1–1.5 mm long, 150–250 µm high, solitary to gregarious, immersed, then raised from host tissue when mature, ellipsoid to irregular, black, coriaceous. Conidiomata wall composed of one layer of dark brown to brown to hyaline cells of textura angularis. Conidiophores mother cells 4.5–6 × 3.5–4.5 µm ( x = 5.4 × 4.3 µm, n = 20), cylindrical, aseptate. Conidiogenous cells 10–48 × 4–5.5 µm ( x = 35.4 × 4.3 µm, n = 20), basauxic, cylindrical, discrete, smooth-walled. Conidia 20–35 × 15–30 µm ( x = 28.5 × 25.6 µm, n = 20), globose to subglobose, dark brown, smooth-walled, with a truncate basal scar.
Culture characters – Ascospores germinating on WA within 24 h and germ tubes developing from the upper cell. Colonies on PDA reaching 50 mm in 1 week at 27°C, in dark, cottony, circular, with irregular edge, white from above and below, becoming dark from the centre after 2 weeks. Mycelium is superficial to immersed in media, with branched, septate, smooth hyphae.
Material examined – China, Yunnan Province, Xishuangbanna Topical Botanical Garden, Bamboo Collection, on bamboo culm, 2 April 2017, Dong-Qin Dai DDQ00403 (HKAS 102408, holotype), ex-type living culture KUMCC 18-0192.
GenBank numbers – ITS: MK070897, LSU: MK070898, SSU: MK070899.
Notes – This species is similar to Arthrinium garethjonesii, which was introduced by Dai et al. (2016) based on morphological and phylogenetic analyses and is only known from the southwest of China. However, A. neogarethjonesii has smaller asci (95–125 × 20–25 μm vs. 125–154 × 35–42 μm) and ascospores (25–30 × 9.5–11 μm vs. 30–42 × 11–16 μm), and the conidia are larger when compared with most Arthrinium species. Based on a Blast search using the ITS sequence in NCBI’s GenBank, the closest hit with the highest similarity is A. garethjonesii (GenBank KY356086; similarity = 575/595 (97%), Gaps = 8/595 (1%)). A similar search using the LSU sequence showed the highest similarity to A. garethjonesii (GenBank KY356091, similarity = 851/854 (99%), no gaps), and the closest hits with A. ovatum (GenBank NG_042782; similarity = 849/875 (97%), no gaps). The phylogenetic tree generated from maximum-parsimony analysis indicated that the new species is close to A. garethjonesii, and these two species cluster in two well-supported clades (MP/BYPP 100/1.00) (Fig. 4).
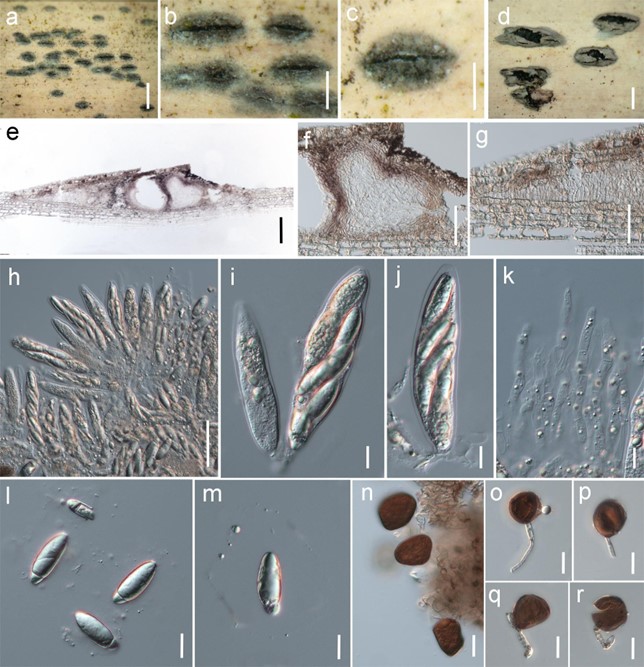
Figure 32 – Arthrinium neogarethjonesii (HKAS 102408, holotype). a-c Appearance of stromata on host. d Appearance of conidiomata on host. e Section of ascostroma. f Section of ascoma. g Stromatic tissue. h-j Asci. k Paraphyses. l, m Ascospores surrounded by a gelatinous sheath. n Conidia. o-r Conidia and conidiogenous cells. Scale bars: a = 1 mm, b-d = 5 µm, e = 100 µm, f-h = 50 µm, i-r = 10 µm.
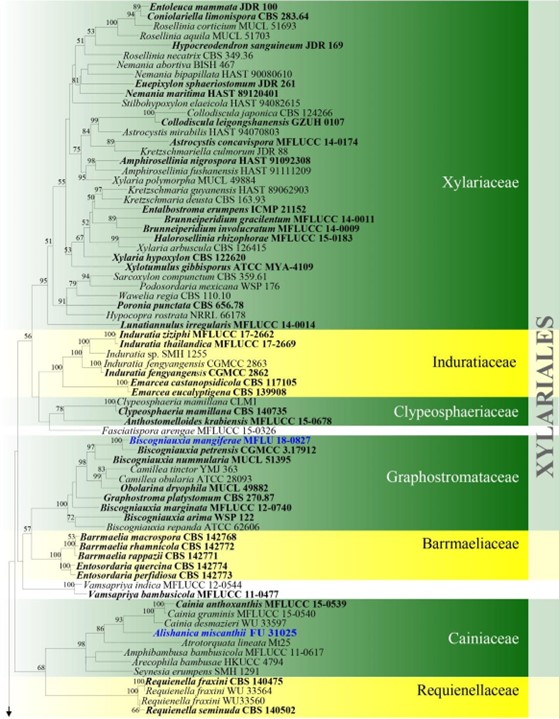
Figure 4 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, rpb2 and tub2 sequence data for Xylariomycetidae. Two hundred and seventy-two strains are included in the combined analyses which comprised 4211 characters (1168 characters for ITS, 937 characters for LSU, 1128 characters for rpb2, 978 characters for tub2) after alignment. Achaetomium macrosporum (CBS 532.94), Chaetomium elatum (CBS 374.66) and Sordaria fimicola (CBS 723.96) are outgroup taxa. Single gene analyses were carried out and the topology of each tree had clade stability. The best RaxML tree with a final likelihood value of – 132297.706952 is presented. Estimated base frequencies were as follows: A = 0.241914, C = 0.251908, G = 0.265558, T = 0.240620; substitution rates AC = 1.281946, AG = 3.512297, AT = 1.499895, CG = 1.121065, CT = 6.472834, GT = 1.000000; gamma distribution shape parameter a = 0.678614. Bootstrap support values for ML greater than 75% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
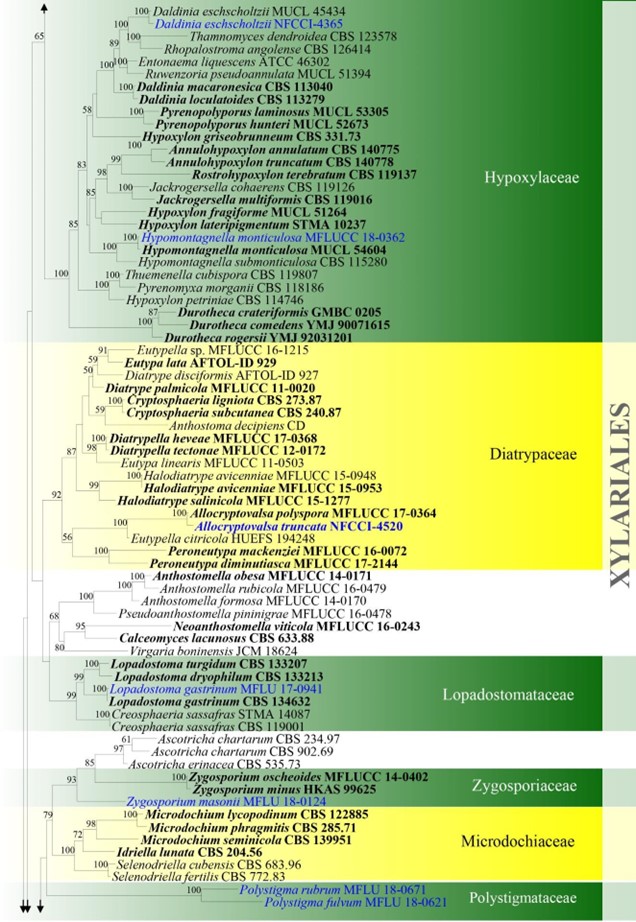
Figure – 4 Continued.
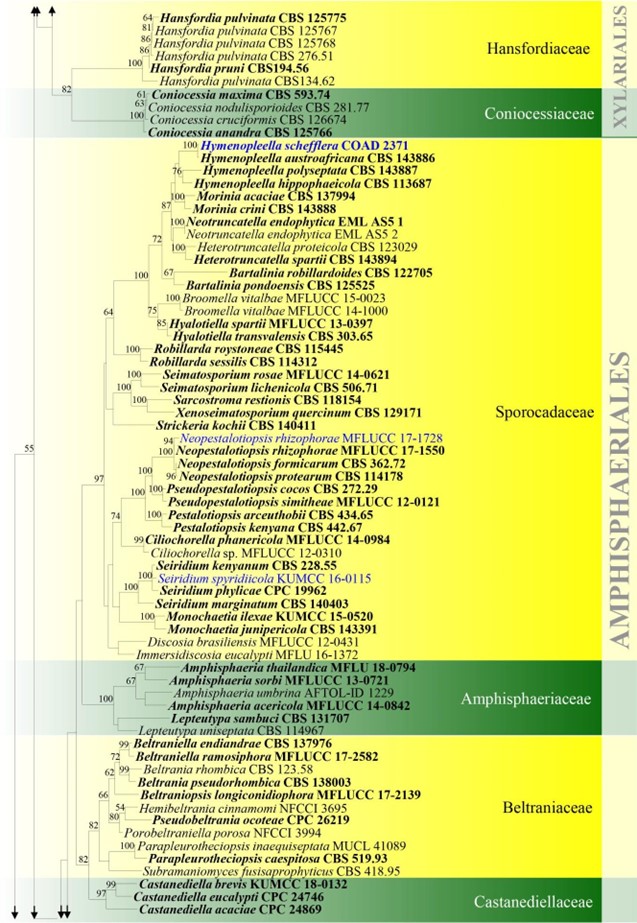
Figure – 4 Continued.

Figure – 4 Continued.
