Alloneottiosporina Nag Raj, Coelomycetous Anamorphs with Appendage-bearing Conidia (Ontario): 121 (1993).
MycoBank number: MB 26427; Index Fungorum number: IF 26427; Facesoffungi number: FoF07107; 3 morphological species (Species Fungorum 2020), 1 species with molecular data.
Parasitic on living leaves of undetermined species of bamboo, grass, and Paspalum distichum. Sexual morph: undetermined. Asexual morph: Conidiomata brown to dark brown or black, pycnidial, separate, gregarious or confluent, immersed to semi-immersed, globose to subglobose, unilocular, glabrous, thick-walled, papillate, ostiolate. Ostiole circular to oval, papillate, centrally located. Conidiomatal wall composed of thick-walled, dark brown to pale brown or hyaline cells of textura prismatica or textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells of two kinds: (a) those producing macroconidia lining the base and side walls of conidiomata, hyaline, annellidic, ampulliform to lageniform, smooth-walled; (b) those producing microconidia restricted to the roof of the conidiomata near the ostiolar channel, hyaline, ampulliform, conical, lageniform or irregular, discrete, without annellations, smooth-walled. Macroconidia hyaline, ellipsoid, clavate, subcylindrical or fusiform, euseptate, constricted at the septa, smooth-walled, bearing mucoid appendages at both ends, apical appendages initially funnel-shaped or widely flared, eventually splitting to become tentaculiform; basal appendage less conspicuous than the apical appendages. Microconidia hyaline, ellipsoid or subglobose to obovate, acute or rounded at the apex, narrow and truncate at the base, smooth-walled.
Type species – Alloneottiosporina carolinensis Nag Raj, Coelomycetous Anamorphs with Appendage-bearing Conidia (Ontario): 122 (1993).
Notes – Alloneottiosporina is characterized by hyaline, ellipsoid, clavate, subcylindrical or fusiform, euseptate conidia bearing tentaculiform or widely flared, mucoid appendages at both ends. Nag Raj (1993) discussed the differences between Alloneottiosporina and other morphologically similar genera, viz., Neottiosporina Subram., Stagonospora (Sacc.) Sacc., Tiarospora Sacc. & Marchal and Tiarosporella Höhn. Two species were accepted in the genus, namely, A. carolinensis and A. infundibularis, but neither has been studied using molecular data. Alloneottiosporina thailandica strain MFLUCC 15–0576 collected from Thailand is described with molecular data. The phylogenetic analysis indicates that Alloneottiosporina may be related to Phaeosphaeriaceae, but in the absence
of ex-type cultures, the phylogenetic position of this genus cannot be confirmed.
Distribution – Australia, USA, Thailand (Nag Raj 1993, this study).
Key to species of Alloneottiosporina
1. Conidia 2–3, unequally septate ……………………………………………………………………………………………………………….. 2
1. Conidia 1–septate…………………………………………………………………………………………………………………. A. thailandica
2. Central cell longer than end cells………………………………………………………………………………………….. A. carolinensis
2. Central cell shorter than end cells……………………………………………………………………………………… A. infundibularis
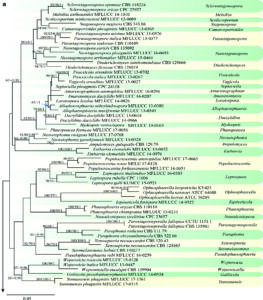
Fig. 1 Phylogenetic tree generated from a maximum likelihood analysis based on a concatenated alignment of LSU, ITS, SSU and tef1 sequences data of Phaeosphaeriaceae. The newly generated nucleotide sequences were compared against the GenBank database via blast search. Related sequences were obtained from (Phookamsak et al. 2017; Zhang et al. 2019) and GenBank. One hundred and six strains are included in the analyses, which comprise 3164 characters including gaps (LSU: 1-834, ITS: 838-1317, SSU: 1326-2269, tef1: 2273-3164). Paraleptosphaeria dryadis CBS 643.86 and Leptosphaeria doliolum CBS 505.75 were used as the outgroup taxa. The tree topology of the maximum likelihood analysis is similar to the maximum parsimony and the Bayesian analysis. The best-scoring RAxML tree with a final optimization likelihood value of − 23616.706146 is presented. The matrix had 961 distinct alignment patterns, with 20.70% of undetermined characters or gaps. Estimated base frequencies were: A = 0.243007, C = 0.236284, G = 0.267159, T = 0.253551; substitution rates AC = 1.136380, AG = 2.921747, AT = 2.414255, CG = 0.638090, CT = 6.696516, GT = 1.000000; gamma distribution shape parameter α = 0.152855. The maximum parsimonious
dataset consisted of constant 2406, 553 parsimony-informative and 205 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 217 steps (CI = 0.295, RI = 0.602, RC = 0.177, HI = 0.705) in the first tree. Maximum parsimony (MPBS) and maximum likelihood (MLBS) bootstrap support values higher than 50%, and Bayesian posterior probabilities ≥ 0.95 (PP) are shown above or below the nodes. Hyphen (“–”) indicates a value lower than 50% for MPBS and MLBS and a posterior probability lower than 0.95 for BYY. The scale bar indicates 0.05 changes. The new isolates are shown in blue
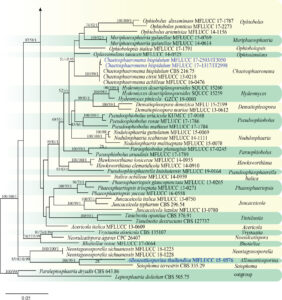
Fig. 1 Phylogenetic tree generated from a maximum likelihood analysis based on a concatenated alignment of LSU, ITS, SSU and tef1 sequences data of Phaeosphaeriaceae. The newly generated nucleotide sequences were compared against the GenBank database via blast search. Related sequences were obtained from (Phookamsak et al. 2017; Zhang et al. 2019) and GenBank. One hundred and six strains are included in the analyses, which comprise 3164 characters including gaps (LSU: 1-834, ITS: 838-1317, SSU: 1326-2269, tef1: 2273-3164). Paraleptosphaeria dryadis CBS 643.86 and Leptosphaeria doliolum CBS 505.75 were used as the outgroup taxa. The tree topology of the maximum likelihood analysis is similar to the maximum parsimony and the Bayesian analysis. The best-scoring RAxML tree with a final optimization likelihood value of − 23616.706146 is presented. The matrix had 961 distinct alignment patterns, with 20.70% of undetermined characters or gaps. Estimated base frequencies were: A = 0.243007, C = 0.236284, G = 0.267159, T = 0.253551; substitution rates AC = 1.136380, AG = 2.921747, AT = 2.414255, CG = 0.638090, CT = 6.696516, GT = 1.000000; gamma distribution shape parameter α = 0.152855. The maximum parsimonious
dataset consisted of constant 2406, 553 parsimony-informative and 205 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 217 steps (CI = 0.295, RI = 0.602, RC = 0.177, HI = 0.705) in the first tree. Maximum parsimony (MPBS) and maximum likelihood (MLBS) bootstrap support values higher than 50%, and Bayesian posterior probabilities ≥ 0.95 (PP) are shown above or below the nodes. Hyphen (“–”) indicates a value lower than 50% for MPBS and MLBS and a posterior probability lower than 0.95 for BYY. The scale bar indicates 0.05 changes. The new isolates are shown in blue
Species
