Neodidymelliopsis salviae Brahmanage, Camporesi & K.D. Hyde, sp. nov.
Index Fungorum number: IF 556375; MycoBank: MB 556365; Facesoffungi number: FoF 06125; Fig. 1
Etymology – Name reflects the host genus.
Holotype – JZBH3470001.
Saprobic on dead and dying twigs and branches of Salvia officinalis. Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 120–375 µm diam (x̅ = 240 µm, n = 5), pycnidial, solitary, scattered, globose, black, semi-immersed to immersed. Pycnidial wall 28–45 μm wide, pseudoparenchymatous, 2–3-layered, composed of oblong to isodiametric cells. Conidiogenous cells phialidic, ampulliform, hyaline. Conidia 6.3–9.3 × 3–4 µm (x̅ = 7.8 × 3.6 µm, n = 40), oblong to ellipsoid, initially hyaline becoming light brown when mature, aseptate occasionally with larger 1-septate conidia, smooth-walled.
Material examined – ITALY, Carpena – Forlì (Province of Forlì-Cesena), on Salvia officinalis (Lamiaceae), 19 March 2018, Camporesi Erio IT 3785 (JZBH3470001, holotype).
GenBank numbers – ITS: MK651827; LSU: MK651828; RPB2: MK736714
Notes – Neodidymelliopsis salvia can be morphologically assigned to Neodidymelliopsis by having pycnidial, conidiomata that are globose, phialidic, hyaline, smooth, ampulliform conidiogenous cells and oblong to ellipsoid, hyaline to pale brown, usually aseptate or occasionally 1-septate conidia (Jayasiri et al. 2017). Phylogenetic analyses based on a combined LSU, ITS, RPB2 and β-TUB sequence data showed that N. salvia and N. sambuci formed a well-supported (85% ML and 1.00 PP) lineage within Neodidymelliopsis (Fig. 2), but morphologically N. sambuci can be distinguished by smaller conidia (4–7 × 2–3.5 µm, Hyde et al. 2019). Based on both morphology and phylogeny, we introduce N. salvia as a new taxon within Neodidymelliopsis
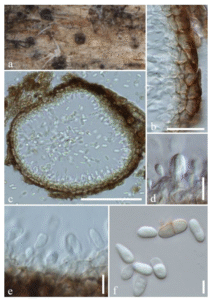
Figure 1 – Neodidymelliopsis salviae (JZBH3470001, holotype). a Submerged conidiomata on host. b Pycnidial wall on host. c Section through conidiomata. d, e Conidiogenous cells and conidial attachments. f Conidia. Scale bars: c = 100, b = 20 µm, d, e = 10, f = 5 µm
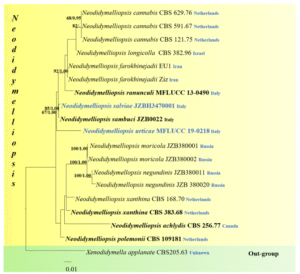
Figure 2 – Maximum likelihood tree based on a combined LSU, ITS, RPB2 and β-TUB sequence dataset. 38 strains of Neodidymelliopsis are included with Xenodidymella applanata (CBS 205.63) as the outgroup taxon. Maximum likelihood bootstrap (ML) values ≥ 65% and Bayesian posterior probabilities (PP) ≥ 0.80% are given above the nodes. The scale bar indicates 0.006 changes. The ex-type strains are in bold and new isolates are in blue
