Asymmetrispora zingiberacearum Tennakoon & C.H. Kuo sp. nov.
MycoBank number: MB; Index Fungorum number: IF; Facesoffungi number: FoF 12711, Fig.
Etymology: The species name reflects the host family Zingiberaceae from which the holotype was collected.
Holotype: MFLU 19-2813
Saprobic on dead stem of Hedychium coronarium (Zingiberaceae). Sexual morph: Undetermined. Asexual morph: Coelomycetous. Conidiomata 100–170 µm high, 120–180 µm diam. (= 140 × 150 µm, n = 10), pycnidial, forming dark brown to black, linear, raised areas on the host surface, semi-immersed to erumpent, solitary or clustered, carbonaceous, globose to sub-globose, ostiolate. Conidiomatal wall 10–20 μm wide, several layers of light brown, thick-walled of textura angularis cells, fusing at the outside indistinguishable from the host tissues. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 5–10 × 3–5 μm (= 8 × 4 µm, n = 20), hyaline, phialidic, cylindrical, slightly tapering towards apex and smooth-walled. Conidia 5–6 × 2–3 (= 5.5 × 2.5 µm, n = 40) μm, aseptate, ellipsoid or obovoid obtuse ends, rarely cylindrical, initially hyaline, light brown at maturity, mostly 2-guttulate, smooth-walled.
Culture characteristics: Colonies on PDA, 15–20 mm diam. after three weeks at 25 °C, colonies from above: medium dense, circular, slightly raised, surface smooth with entire edge, velvety, with smooth aspects, white to cream at the margin, grey in the centre; reverse: yellowish brown at the margin, light brown in the centre, mycelium white to whitish cream or grey.
Material examined: Taiwan, Chiayi, Ali Shan Mountain, Fanlu Township area, Dahu forest, on dead stem of Hedychium coronarium (Zingiberaceae), 25 September 2018, D. S. Tennakoon, ROD016A (MFLU 19-2813, holotype); ex-type living culture, NCYUCC 19-0273. ibid. 28 September 2018, ROD016B (NCYU 19-0115, paratype); ex-paratype living culture, NCYUCC 19-0283.
Notes: According to the multi-gene phylogenetic analyses (LSU, SSU, ITS and tef1-α), our strains (MFLU 19-2813 and NCYU 19-0115) grouped within Asymmetrispora isolates (A. tennesseensis and A. mariae) in a strongly supported clade (100% ML, 1.00 BYPP, Fig. 4). Both A. tennesseensis and A. mariae species have been recorded as their sexual morphs. Thus, we could not compare the morphological differences between our new collection and A. tennesseensis and A. mariae. Therefore, we compared the ITS (+5.8S) and tef1-α -α gene regions base pair differences. There are 10 base pair differences (1.96%) across 510 nucleotides across the ITS (+5.8S) gene region and 26 base pair differences (3.59%) across 724 nucleotides across the tef1-α gene region between our collection (MFLU 19-2813) and A. mariae (CBS 140732). In addition, there are 29 base pair differences (3.86%) across 724 nucleotides across the tef1-α gene region between our collection (MFLU 19-2813) and A. tennesseensis (ANM 911). Therefore, we introduce our collection as a new species, A. zingiberacearum from dead stems of H. coronarium (Zingiberaceae). It will be interesting to expand this genus with further collections to resolve the sexual and asexual connection of Asymmetrispora species.
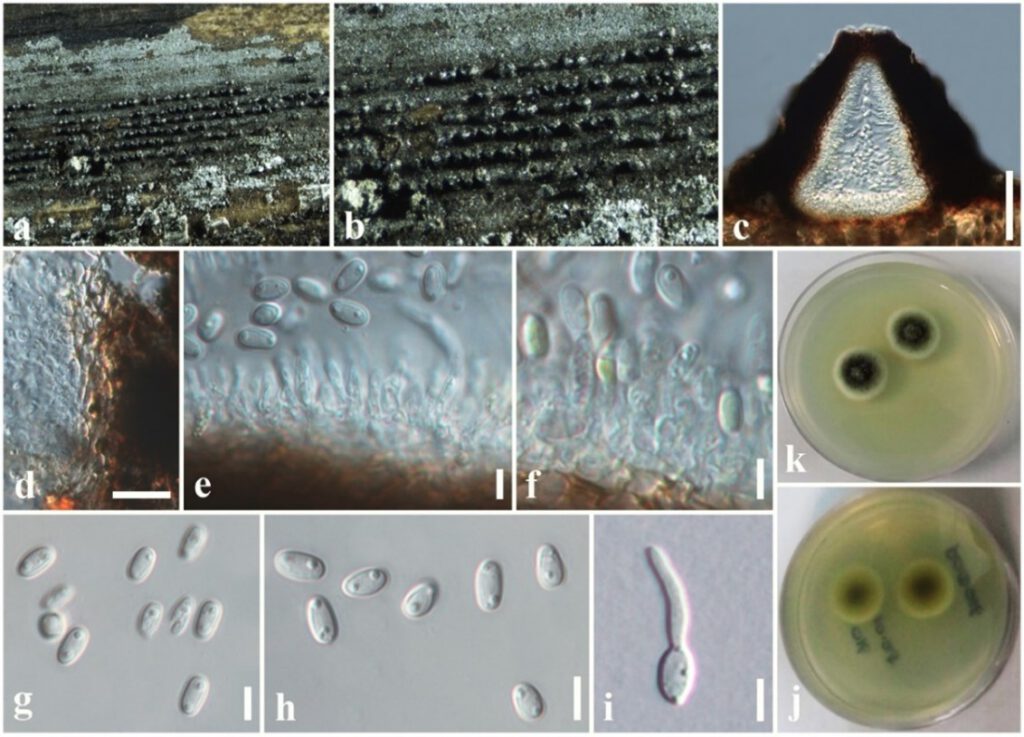
Figure *. Asymmetrispora zingiberacearum (MFLU 19-2813, holotype). (a, b) Conidiomata on the host. (c) Section through conidioma. (d) Conidiomatal wall. (e, f) Conidiogenous cells and developing conidia. (g, h) Conidia. (i) A germinated conidium. (j) Colony from below (on PDA). (n) Colony from above (on PDA). Scale bars: (c) = 50 µm, (d) = 10 µm, (e–i) = 5 µm.
