Eriomyces heveae Huanraluek, Thambugala & K.D. Hyde,sp. nov.
MycoBank number: MB 556539; Index Fungorum number: IF 556539; Facesoffungi number: FoF 06152; Fig. 92
Etymology: The species epithet "heveae" refers to the host genus Hevea on which holotype occurs.
Holotype: MFLU 18-0758
Saprobic on dead twigs of Hevea brasiliensis Müll. Arg. Sexual morph Ascomata 240–380(–445) μm high × 215–325 μm diam., immersed beneath a raised dark brown to black pseudoclypeus, papilla erumpent through host surface, solitary or aggregated, scattered, black, coria- ceous, unilocular, subglobose to flask-shaped, or conical, with a raised neck. Ostiole central, papillate, periphysate. Peridium 30–56 μm wide, comprising several layers of heavily pigmented to hyaline, thick-walled, cells of textura epidermoidea, fusing at the outside with the host cells. Hamathecium composed of 1–3 μm wide, numerous, rarely septate, branched or unbranched, cellular pseudoparaphyses. Asci 52–102 × 9–13 μm ( x̄ = 80 × 10.2 μm, n = 20), poly-spored, bitunicate, cylindrical to cylindric-clavate, with a long to short, narrow, thin-walled, furcate pedicel, apically broadly rounded. Ascospores 4.5–6.5 × 1–2 μm ( x̄ = 5.5 × 1.4 μm, n = 50), crowded, ellipsoidal, aseptate, straight to slightly curved, hyaline, smooth-walled, with bipolar guttules. Asexual morph Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes produced from one end. Colonies growing on PDA reaching 25 mm diam. after 12 days at 25 °C, circular, falt, slightly dense, surface white to iron- grey, reverse brownish to smoke-grey, white at the margin, smooth surface with entire to slightly curled edge.
Material examined: THAILAND, Sukhothai Province, Si Satchanalai District, on dead twigs of Hevea brasilien- sis (Euphorbiaceae), 2 January 2017, N. Huanraluek, Sh1 (MFLU 18-0758, holotype); ibid., KUN-HKAS 97497, iso- type, ex-type living culture MFLUCC 17-2232.
GenBank numbers: ITS =MH118116, LSU = MH109524.
Notes: – Based on a megablast search of NCBIs Gen- Bank nucleotide database using LSU sequences of strain MFLUCC 17-2232, the closest hit was Pseudopassalora gouriqua (CPC 1811; 95.47% similarity), followed by Heleiosa barbatula (JK 5548I; 95.70% similarity). In the present phylogeny (Fig. 93), Eriomyces heveae shows a close affinity to Heleiosa barbatula. Nevertheless, Eriomyces heveae differs from Heleiosa barbatula in having poly-spored asci and hyaline, oblong to allantoid, aseptate ascospores (Kohlmeyer et al. 1996). In addition, H. barbatula was described from Juncus roemerianus Scheele (Juncaceae) in salt marshes of the USA east coast, while Eriomyces heveae was reported from Hevea brasiliensis in Thailand. Therefore, considering the distinct morphological traits and phylogeny E. heveae is introduced as a new species. Ascospores of Eriomyces morphologically resemble those of Cytospora Ehrenb. and Eutypella (Nitschke) Sacc. in Sordariomycetes.
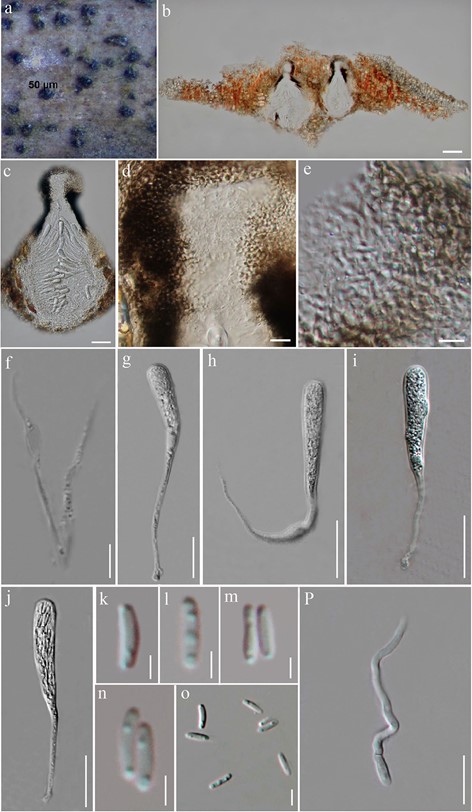
Fig. 92 Eriomyces heveae (MFLU 18-0758, holotype). a Appearance of ascomata on host surface. b, c Vertical sections through ascomata. d Vertical sections through the apex of ascoma. e Peridium. f Pseudoparaphyses. g–j Asci. k–o Ascospores p Germinating ascospore. Scale bars: b, c = 50 µm, d, e = 25 µm, f–j = 20 µm, p = 10 µm, k–o = 5 µm
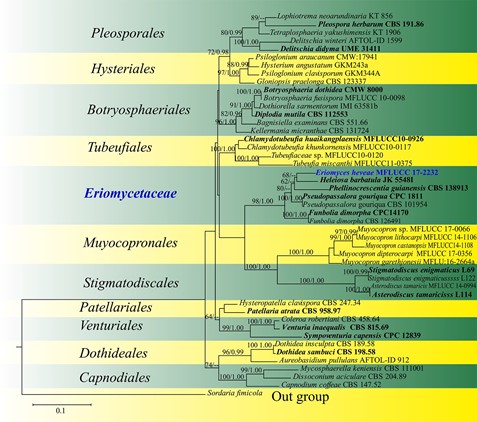
Fig. 93 Phylogram generated from maximum likelihood analysis based on LSU sequence data representing Eriomyces (Dothideomyetes, families incertae sedis) and the closely related orders in Dothideomyetes. Related sequences are taken from Suetrong et al. (2009). Forty-seven strains are included in the combined analyses which comprise 886 characters after alignment. Sordaria fimicola (CBS 508.50) (Sordariomycetes) is used as the outgroup taxon. The best RAxML tree with a final likelihood values of − 7924.174629 is presented. Estimated base frequencies were as follows: A = 0.245455, C = 0.227992, G = 0.318413, T = 0.208140; substitution rates AC = 0.803611, AG = 2.255570, AT = 0.841920, CG = 1.068389, CT = 8.568242, GT = 1.000000. Tree topology of the maximum like- lihood analysis is similar to the Bayesian analysis. Bootstrap values for maximum likelihood (ML) equal to or greater than 70% and clade credibility values greater than 0.90 (the rounding of values to 2 decimal proportions) from Bayesian-inference analysis labeled on the nodes. Ex-type strains are in bold and black, the new isolate is indicated in bold and blue
