Trochilispora VP Abreu, AWC Rosado & OL Pereira, in Hyde et al., Fungal Diversity: 10.1007/s13225-019-00429-2, [169] (2019)
MycoBank number: MB 555484; Index Fungorum number: IF 555484; Facesoffungi number: FoF 04859
Etymology – From the family Trochilidae (hummingbirds) and –spora.
Type species – Trochilispora schefflerae Abreu VP, Rosado AWC & Pereira OL
Associated with leaf spot disease on Schefflera morototoni. Asexual morph Conidiomata acervular, epiphyllous, scattered and occasionally confluent, subepidermal in origin, erumpent, rounded to oval in outline, unilocular, brown or black, basal stroma thick, of textura angularis, cells thick-walled and almost colourless; lateral walls thick, cells thick-walled, pale brown to brown. Conidiophores cylindrical to subcylindrical, formed in the concavity of the conidioma, unbranched, hyaline, smooth-walled. Conidiogenous cells discrete, annellidic with 2 annellations, hyaline, thin- and smooth-walled. Conidia fusiform, straight or slightly curved, 3–4-septate, concolourous, smooth, bearing apical appendage tubular, filiform, single, not centric, unbranched and basal appendage absent; basal cell hyaline to subhyaline, obconic to conic, smooth and thin-walled; 2–3 median cells doliiform, smooth, concolourous, brown, septa darker than the rest of the cell; apical cell hyaline to subhyaline, subconical to hemisphaerical, thin- and smooth-walled. Sexual morph Undetermined.
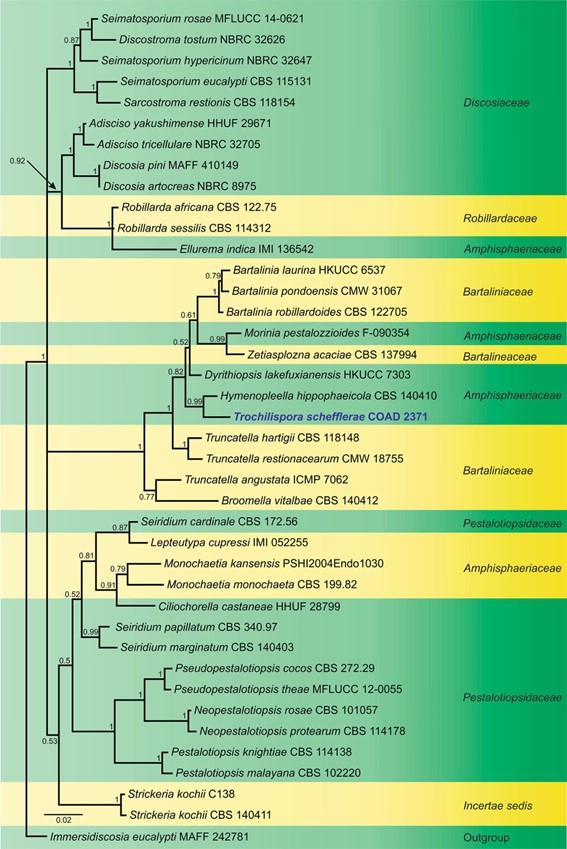
Fig. 1 Phylogram generated from Bayesian Inference analysis based on combined ITS and LSU sequence data for several closely related genera in Amphisphaeriales. Sequence data of ex-type or ex- epitype cultures are taken from Senanayake et al. (2015), and the closest hits of the GenBank database were included in this study. The combined genes sequence analysis included 40 taxa, which comprise 1405 characters (591 characters for ITS, 814 characters for LSU), and outgroup taxon Immersidiscosia eucalypti (MAFF 242781). Bayesian posterior probability is indicated at the nodes, and values equal to or greater than 0.95 are in bold. Isolate numbers are indicated after species names. The ex-type or ex-epitype strains are in bold and black. The newly generated sequence is indicated in bold and blue

Fig. 2. Phylogram of maximum likelihood analysis based on ITS, TUB2 and TEF1-a sequences for Pestalotiopsis species. Related sequences were obtained from Maharachchikumbura et al. (2012, 2014). Neopestalotiopsis saprophyta (MFLUCC 12-0282) is used as the outgroup taxon. Sixty-five strains are included in the combined analyses, which comprise 1545 characters (565 characters for ITS, 472 characters for TUB, and 508 characters for TEF1-a) after alignment. Single gene analyses are carried out, and the topology of each tree had clade stability. The tree topology of the maximum likelihood analysis is similar to the maximum parsimony analysis. The best RaxML tree with a final likelihood value of – 10917.490545 is presented. The matrix had 716 distinct alignment patterns, with 12.01% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.234122, C = 0.292180, G = 0.213547, T = 0.260152; substitution rates AC = 1.238816, AG = 3.435200, AT = 1.254551, CG = 1.048645, CT = 4.518698, GT = 1.000000; gamma distribution shape parameter a = 0.803705. Maximum parsimony analysis of 945 constant characters and 384 informative characters resulted in two equally most parsimonious trees of 1643 steps (CI = 0.525, RI = 0.685, RC = 0.360, HI = 0.475). Bootstrap support values for ML (first set) and MP equal to or greater than 50% are given above the nodes. The ex-type strains are in bold and black. The newly generated sequence is indicated in bold and blue
Species
