Tirisporellales Suetrong, E.B.G. Jones & K.L. Pang, Fungal Divers. 73(1): 42 (2015)
MycoBank number: MB 812597; Index Fungorum number: IF 812597; Facesoffungi number: FoF 00860;
The monotypic order Tirisporellales is placed in the class Sordariomycetes, subclass Diaporthomycetidae and comprises a single family Tirisporellaceae. Suetrong et al. (2015) introduced Tirisporellaceae with two monotypic genera Tirisporella and Thailandiomyces. Jones et al. (2015) introduced Tirisporellales to accommodate the genera Tirisporella, Thailandiomyces with an additional genus Bacusphaeria. In our phylogram generated with concatenated LSU and SSU sequence data, the genera Tirisporella (100% ML, 100% MP), Thailandiomyces (93% ML, 100% MP), and Bacusphaeria (100% ML, 100% MP) formed strongly supported clades within Tirisporellales (Fig. 25). In Fig. 1 this order formed an internal clade of Diaporthales and this observation is supported in Hongsanan et al. (2017). However, Tirisporellales was not formally synonymized under Diaporthales by either Hyde et al. (2017a) or Hongsanan et al. (2017). Tirisporellaceae is primarily aquatic with both freshwater and marine taxa with a hyphomycetous asexual morphs which are not usually found in Diaporthales (Zhang et al. 2019). Therefore, we do not place this order in Diaporthales. The divergence time for Tirisporellales is estimated as 142 MYA (Fig. 2). Currently there is one family and three genera in this order (this paper).
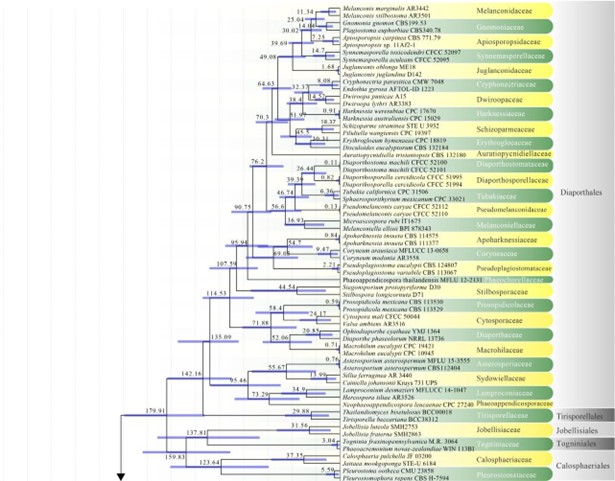
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
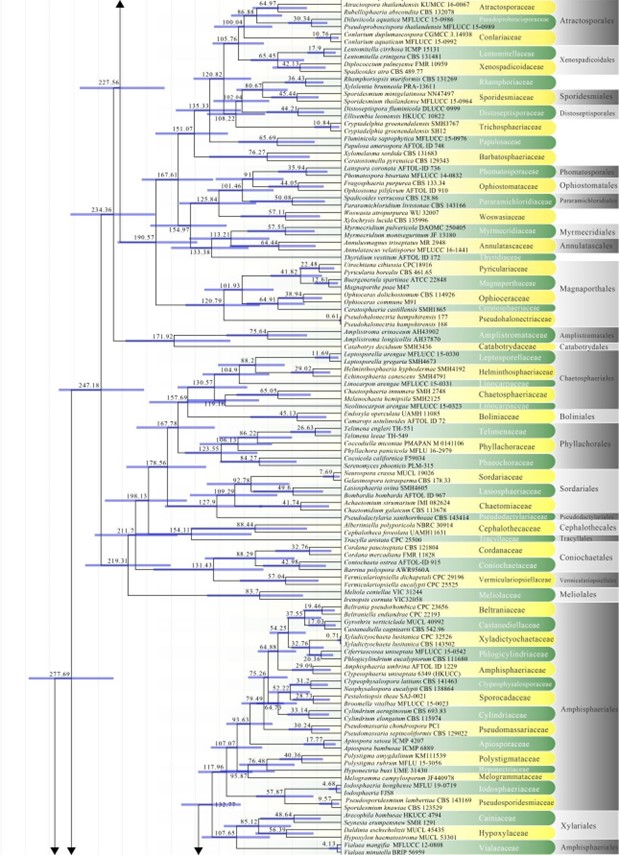
Figure 2 – Continued.
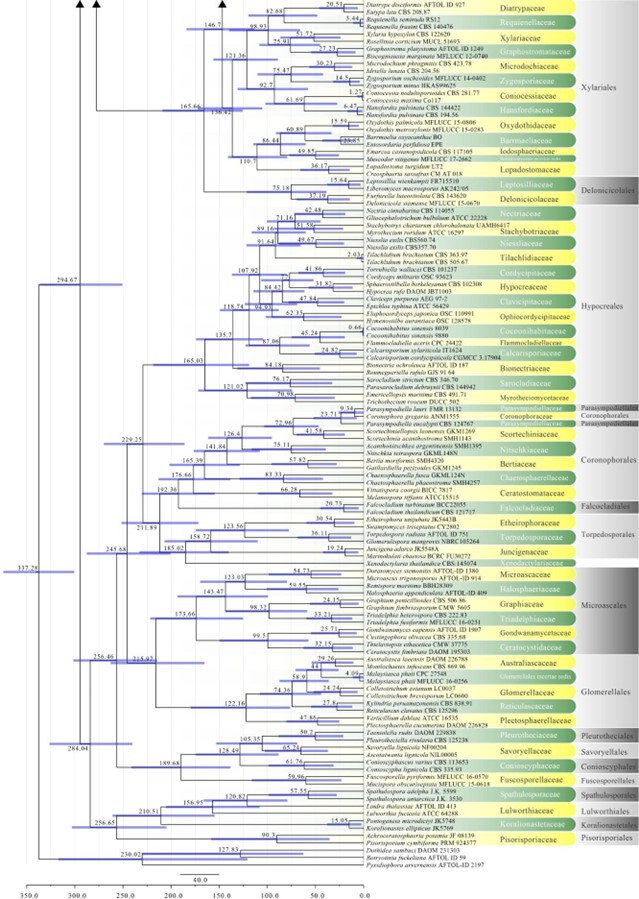
Figure 2 – Continued.
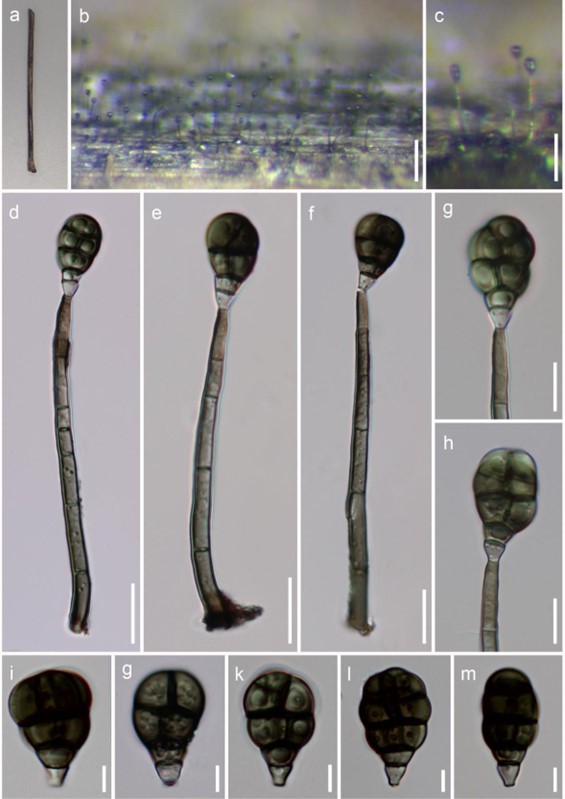
Figure 25 – Acrodictys aquatica (Material examined – THAILAND, Chiang Rai Province, Muang, Ban Nang Lae Nai, on decaying wood submerged in a freshwater stream, 31 December 2016, NG Liu, CR004, MFLU 18-0040, holotype). a Specimen. b, c Colonies on substrate. d-f Conidiophores and conidia. g, h Conidiogenous cells and conidia. i-m Conidia. Scale bars: b = 100 μm, c = 50 μm, d-f = 15 μm, g, h = 10 μm, i-m = 5 μm.
Families
