Savoryellomycetidae Hongsanan, K.D. Hyde & Maharachch., Fungal Divers. 84: 35 (2017)
MycoBank number: MB 553214; Index Fungorum number: IF 553214; Faceoffungi number: FoF 03354;
According to Maharachchikumbura et al. (2016b) and Yang et al. (2016b) Conioscyphales was assigned to Diaporthomycetidae, genera incertae sedis, while Fuscosporellales, Pleurotheciales, and Savoryellales were included in the subclass Hypocreomycetidae. In the phylogenetic and molecular clock analyses of (Hongsanan et al. 2017, Hyde et al. 2017a), Conioscyphales, Fuscosporellales, Pleurotheciales, and Savoryellales clustered together as a distinct clade, with a stem age of 268 MYA. Hence, they were referred to a new subclass Savoryellomycetidae by Hongsanan et al. (2017) and this was reinforced in the paper by Dayarathne et al. (2019a). Our phylogenetic analyses with combined LSU, SSU, ITS and rpb2 sequence data also showed Conioscyphales, Fuscosporellales, Pleurotheciales, and Savoryellales formed well-supported distinct clades (100% ML, 1.00 PP, 100% ML, 1.00 PP, 98% ML, 1.00 PP and 100% ML, 1.00 PP, respectively) within the subclass Savoryellomycetidae (Fig. 10). Currently there are four orders and four families in this subclass.
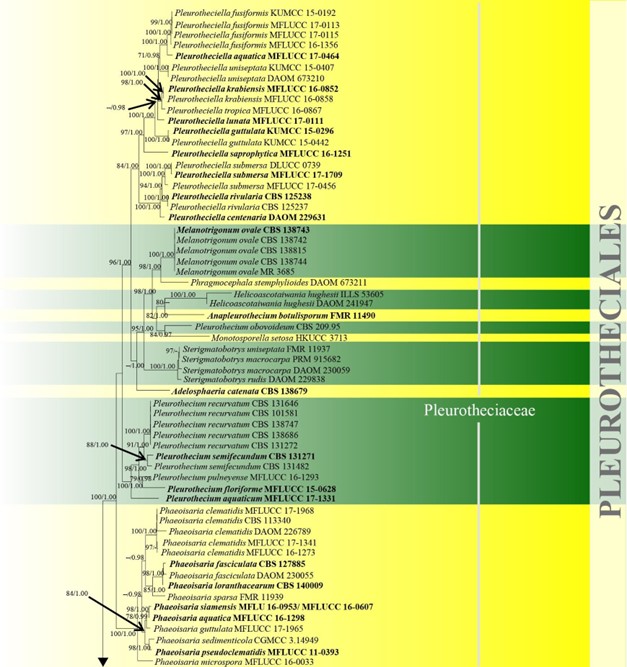
Figure 10 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and rpb2 sequence data of Conioscyphales, Fuscosporellales, Parasympodiellales, Pleurotheciales and Savoryellales. One hundred and twenty-nine strains are included in the combined analyses which comprised 4253 characters (910 characters for LSU, 1499 characters for SSU, 759 characters for ITS and 1085 characters for rpb2) after alignment. Lindra thalassiae (AFTOL-ID 413) and Lulworthia uniseptata (AFTOL-ID 5014) are used as outgroup taxa. Single gene analyses and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -50774.805453 is presented. Estimated base frequencies were as follows: A = 0.233897, C = 0.263834, G = 0.294235, T = 0.208033; substitution rates AC = 1.342378, AG = 2.736022, AT = 1.339928, CG = 0.989078, CT = 6.301203, GT = 1.000000; gamma distribution shape parameter a = 0.440908. Bootstrap support values for ML greater than 70% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. The tree is rooted with Lindra thalassiae (AFTOL-ID 413) and Lulworthia uniseptata (AFTOL-ID 5014). Type strains are in bold and black. The newly generated sequence is indicated in blue.
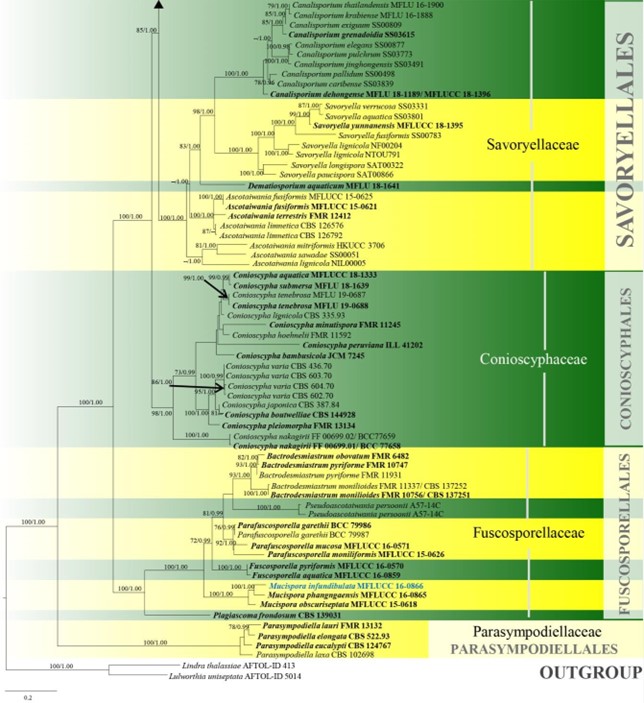
Figure 10 – Continued.
