Pseudohelminthosporium clematidis Phukhams. & K.D. Hyde, sp. nov.
MycoBank number: MB 557192; Index Fungorum number: IF 557192 ; Facesoffungi number: FoF 07284; Fig. 37.
Etymology: The epithet “clematidis” refers to the host substrate.
Holotype: MFLU 17–1494.
Saprobic on decaying wood or herbaceous plant mate- rial in terrestrial habitats. Sexual morph: Undetermined. Asexual morph: Colonies on Clematis sikkimensis effuse, black, hairy, scattered, dark brown. Mycelium immersed, on the substrate surface forming stroma-like aggregations of dark brown sheet. Conidiophores 125–435 × 9–20 μm ( x̄ = 230 × 12 μm, n = 20), macronematous, simple, solitary, branched at the apex, stipes straight or flexuous, cylindrical, erect, septate, smooth, dark brown to reddish brown, 8–14 septa, brown, well-defined small pores with dark scar at the apex, smooth or verruculose. Conidiogenous cells 17–60 × 9–12 μm ( x̄ = 40 × 12 μm, n = 10), monotretic or polytretic, integrated, terminal, becoming intercalary, dolii- form to oblong, pale brown. Conidia 63–142 × 16–26 μm ( x̄ = 87 × 20 μm, n = 20), phragmosporous, acrogenous, broad fusiform or obclavate, distoseptate at the early state, 3–5-euseptate at maturity, slightly constricted at septa, dark brown to reddish brown, verrucose, gradually tapering to 13 μm at the distal end, with a dark brown to black scar present at the base, subhyaline, elongate cells at the upper end of conidia, with (1–)2 guttules in each cell.
Culture characters: Colonies on MEA reaching 40 mm diam. after 4 weeks at 25 °C. Culture from above, brownish grey, dark green, white in the center, forming cream fluffy mycelium at the edge, dense, umbonate, raised with concave edge, rough, dull, lobate, radially furrowed, with brown pigment slightly diffusing into the agar; reverse dark brown.
Material examined: Thailand, Chiang Rai Province, Mae Sai District, dead stems of Clematis sikkimensis, 24 June 2017, C. Phukhamsakda & M. van de Bult, CMTHDT02 (MFLU 17–1494, holotype); ex-type living culture, MFLUCC 17–2086.
Host: Clematis sikkimensis—(This study).
Distribution: Thailand—(This study).
GenBank accession numbers: LSU: MT214567; SSU: MT226683; ITS: MT310612; tef1: MT394627; rpb2: MT394690.
Notes: In a BLASTn search of GenBank, the closest match for the LSU sequence of MFLUCC 17–2086 was Preussia terricola strain CBS 317.65 (GQ203725) with 96.77% similarity. The closest match for the ITS region was Forliomyces uniseptata strain MFLUCC 15–0765 (NR_154006). Based on the multi-locus phylogenetic support (Fig. 2), Pseudohelminthosporium formed a separate lineage related to Neomassarina species but lacked back- bone support. Therefore, Pseudohelminthosporium is treated as a distinct genus in Neomassarinaceae.
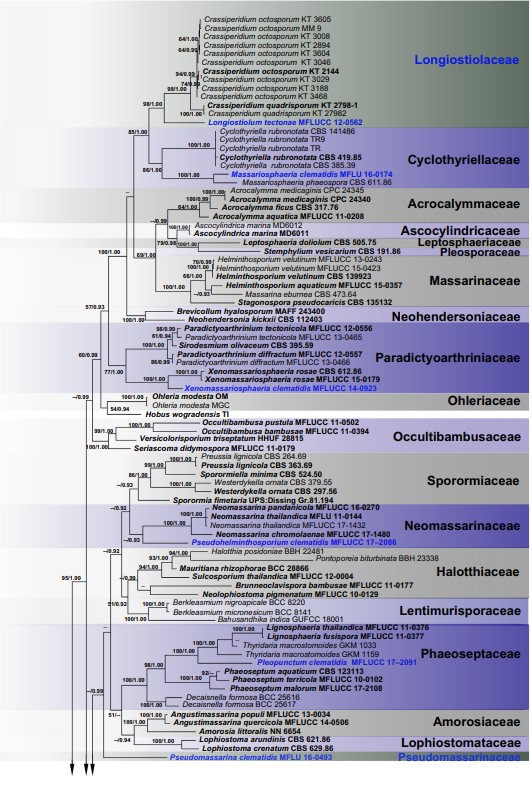
Fig. 2 The Bayesian 50% majority-rule consensus phylogram based ▸ on combined LSU, SSU, ITS, tef1 and rpb2 sequence data of related families in Pleosporales. The topology and clade stability of the combined gene analyses was compared to the single gene analyses. The tree is rooted with species of Hysteriales. One hundred and fifty- three strains were included in the DNA analyses which comprised 4394 characters (848 characters for LSU, 1044 characters for SSU, 556 characters for ITS, 910 characters for tef1, and 1036 characters for rpb2, including gap regions). The tree from the maximum likelihood analysis had similar topology to the Bayesian analyses. The best scoring RAxML tree had a final likelihood value of − 73089.933914. The matrix had 2676 distinct alignment patterns, with 40.81% undetermined characters and gaps. Estimated base frequencies were as follows; A = 0.246412, C = 0.245743, G = 0.272077, T = 0.235768; substitution rates AC = 1.617324, AG = 3.695355, AT = 1.662826, CG = 1.183453, CT = 8.283938, GT = 1.000000; gamma distribution shape parameter α = 0.635095. The GTR + I + G model was selected for every partition in Bayesian analysis. Bootstrap values (BS) greater than 50% BS (ML, left) and Bayesian posterior probabilities (BYPP, right) greater than 0.90 are given at the nodes. Hyphens (-) represent support values less than 50% BS/0.90 BYPP. The type sequences are in bold and the species determined in this study are indicated in blue
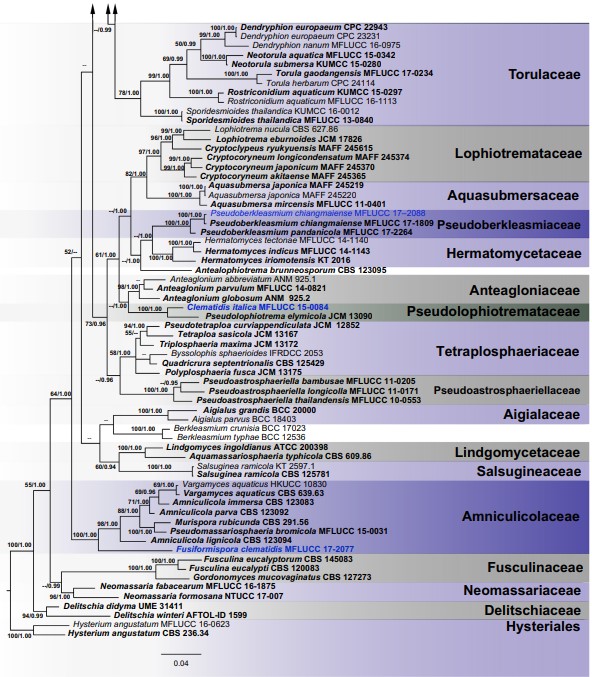
Fig. 2 (continued)
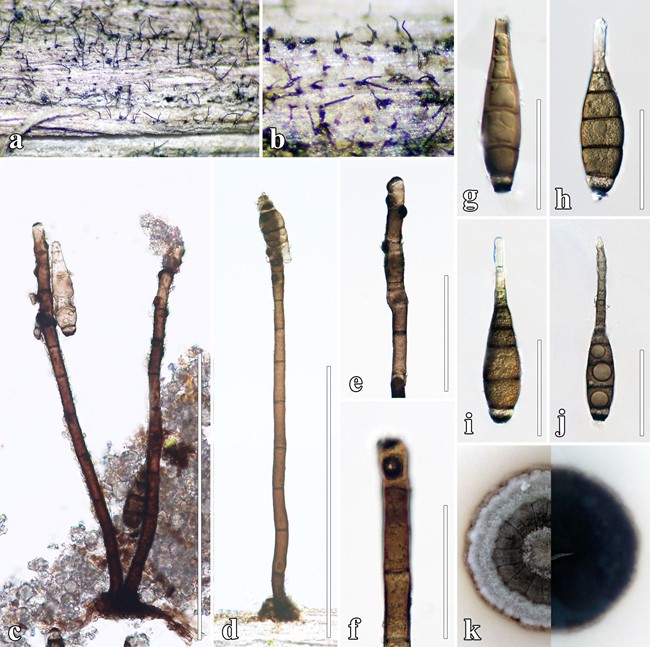
Fig. 37 Pseudohelminthosporium clematidis (MFLU 17–1494, hol- otype). a, b Conidiophores on natural substrate (Clematis sikkimensis). c, d Mononematous conidiophores. e, f Conidiogenous cells and conidia. g–j Conidia (Black basal conidia). k Culture characteristics on MEA. Scale bars: c, d = 500 μm, e = 100 μm, f–j = 50 μm
