Pseudocercospora vignae Y.J. Chen & K.D. Hyde, sp. nov.
MycoBank number:MB 558824; Index Fungorum number: IF 558824; Facesoffungi number: FoF 10530; Fig. 3
Etymology: Refers to the host, Vigna sp.
Holotype: MFLU 21-0132
Pathogenic on leaves of Vigna sp. Leaf spots showing typical lesion with distinct chlorotic margins scattered over leaves, amphigenous, irregular to angular, pale to dark brown becoming darker with age. The lesions are surrounded by yellow halo. Sexual morph: Not observed. Asexual morph on host. Stromata (x̅ = 40 μm, n = 15), erumpent, pale olivaceous brown. Conidiophores 4–20 × 2.6–5.5 µm (x̅ = 10 × 3.8 µm, n = 15) numerous, fasciculate, grouped in dense fascicles, unbranched, smooth, subcylindrical, straight to curved, 0−2-septa, pale brown to hyaline. Conidiogenous cells 9–24 × 2–4 µm (x̅ = 13.8 × 3 µm) unbranched, terminal, integrated, smooth, un-thickened, tapering to a truncated apex, pale brown. Conidiogenous loci, inconspicuous, un-thickened and not darkened. Conidia 22–105 × 2.3–3.6 µm (x̅ = 59 × 3 µm, n = 40) solitary, holoblastic, straight or slightly curved, smooth, thin-walled, obclavate-cylindrical, both ends rounded to long obconic-truncate, 1–7 septa, subhyaline to pale olivaceous-brown. Hila un-thickened, not darkened.
Culture characteristics: On PDA reaching a diam. of 0.5 cm in 7 days at 25 °C, edge irregular, umbonate, margin undulate, greenish-grey to dark grey from above with pale grey in the center, dark grey to black from below.
Material examined: THAILAND, Chiang Rai, Thasud, Muang, on leaf of Vigna sp., 31 Jul 2019, Y.J. Chen, MFLU 21-0132 holotype, ex-type living culture, T19-1479.
GenBank accession numbers ITS: XXX tef: XXX act: XXX
Notes: The new taxon Pseudocercospora vignae formed a sister clade to P. beijingensis and P. pyracanthigena with statistical support (Fig. 4). Pseudocercospora vignae shows 18 bp difference in ITS, 38 bp in tef, and 12 bp in act with P. beijingensis. Pseudocercospora vignae differs from P. beijingensis by its longer conidia (P. vignae: 22−105 µm vs. P. beijingensis: 13–72 µm), and smaller stromata (40 vs. 100 µm) (Wang et al. 2019b). Pseudocercospora vignae is distinguishable from P. pyracanthigena by having longer and wider conidiophores (4–20 × 2.6–5.5 µm vs. 7–15 × 2–3 µm) and more conidial septa (1−7 vs. 1−4-septate) (Crous et al. 2013). Besides, on the plant genus Vigna, Pseudocercospora vignae-reticulatae, P. vignicola and P. vignigena are hitherto known but lack of molecular data. Newly described taxon differs from P. vignae-reticulatae by its shorter conidiophores (P. vignae: 4–20 µm vs. P. vignae-reticulatae: 40–250 μm), narrower conidia (P. vignae: 2.3−3.6 µm vs. P. vignae-reticulatae: 4–6.5 μm), and less conidial septa (1−7 vs. 1−12-septate). Pseudocercospora vignae differs from P. vignicola and P. vignigena by shorter conidiophores (P. vignae: 4–20 µm vs. P. vignicola: 22–75 μm vs. P. vignigena: 22–75 μm), and longer conidia (P. vignae: 22–105 µm vs. P. vignae: 30–60 µm vs. P. vignigena: 33–60 μm).

Fig. 3 Pseudocercospora vignae (MFLU 21-0132, holotype). a Host. b, c Leaf spots on host. d Conidiomata on host substrate. e–g Conidiogenous cells and conidiophores. h–i Immature conidia. j–o Conidia. p Germinated conidium. q Colony. Scale bars c =1.5 mm, d = 800 μm, e = 25 μm, f–i = 15 μm, j–o = 20 μm, p = 25 μm.
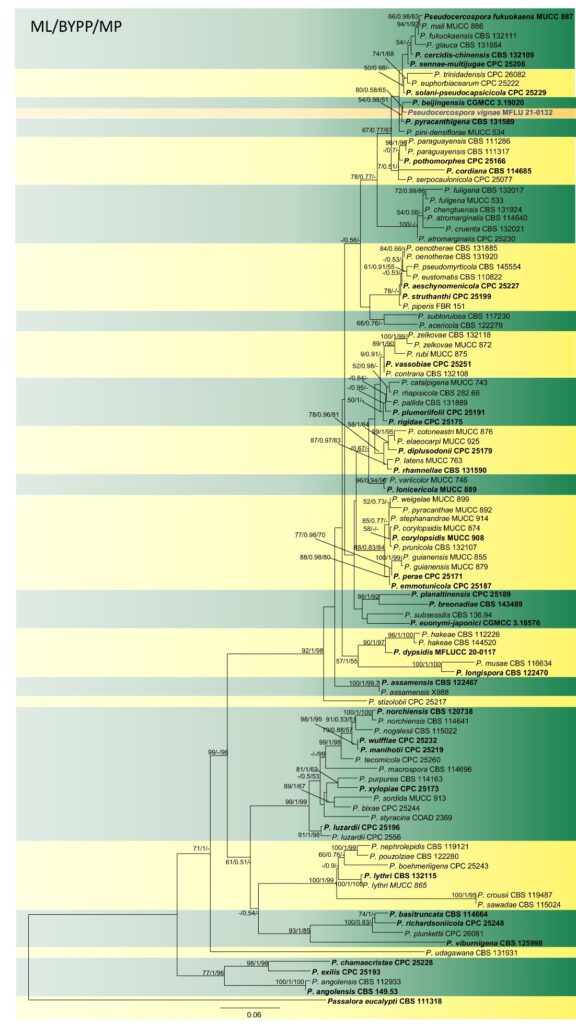
Fig. 4 Phylogram generated from maximum likelihood analysis based on combined ITS, tef and act sequence data of Pseudocercospora species. Related sequences were retrieved from GenBank and 103 strains were included in the analysis of the combined loci which comprised 1161 characters after alignment. The tree is rooted with Passalora eucalypti (CBS 111318). The best scoring RAxML tree had a final likelihood value of -12533.711659 . Estimated base frequencies were: A = 0.221839, C = 0.280126, G = 0.249394, T = 0.248641; substitution rates AC = 1.401581, AG = 3.305591, AT = 1.474641, CG = 0.775502, CT = 4.708205, GT = 1.000000. In our analysis, GTR-gamma model was used for each partition in Bayesian posterior analysis. Maximum parsimony analysis of 403 parsimony informative characters resulted in a most parsimonious tree (CI = 0.420, RI = 0.775, RC = 0.326, HI = 0.580). Bootstrap values (BS) from maximum likelihood, maximum parsimony higher than 50% BS and Bayesian posterior probabilities greater than 0.50 are given at the nodes. Hyphens (-) represent support values less than 50% BS/0.50 BYPP. The ex-type strains are in bold and black. The newly generated sequence is in bold and blue.
