Podosporaceae X. Wei Wang & Houbraken, Stud. Mycol. (2019)
MycoBank number: MB 829841; Index Fungorum number: IF 829841; Facesoffungi number: FoF 06877; 92 species.
Saprobic on dung. Sexual morph: Ascomata superficial to immersed in medium, solitary or loosely aggregated, ostiolate and ovoid to obpyriform, or lacking ostioles, globose to subglobose, glabrous or having hypha-like to seta-like hairs. Peridium membranaceous to coriaceous, usually opaque, in some species semi-translucent. Asci (2–)4- or 8- or multi-spored, unitunicate, cylindrical to elongated clavate or fusiform, pedicellate, with or without an apical thickened ring, evanescent or persistent until ascospores mature. Ascospores 1–2-seriate, 1-celled and pigmented, or 2-celled and composed of a larger, pigmented upper cell and a smaller, pale or hyaline cell, with or without appendage, usually smooth, in a few species ornamented. Asexual morph: Hyphomycetous. not observed or cladorrhinum-like: Conidiophores micronematous, reduced to conidiogenous cells. Conidiogenous cells intercalary or occasionally terminal, originating lateral or terminal peg-like structure with a flaring collarette, with blastic conidia. Conidia single-celled, hyaline, smooth, usually with a truncated base and a rounded apex (adapted from Wang et al. 2019a).
Type genus – Podospora Ces.
Notes – Podosporaceae was invalidly introduced by Hochberzanke (1930). The proposed Podosporaceae introduced by Wang et al. (2019a) is based on Podospora, but they are delimited based on morphology and DNA sequence data. Podosporaceae is sister to Chaetomiaceae in Sordariales and accommodates Podospora, Trangularia and Cladorrhinum, which were positioned in the polyphyletic family Lasiosphaeriaceae (Wang et al. 2019a). Based on the available sequences of Apiosordaria, we consider this genus should be placed in Podosporaceae (Fig. 23). However, Mouchacca & Gams (1993) indicated that several Apiosordaria species have asexual morph connections with Cladorrhinum. Further work is required to determine the additional taxa in this family.
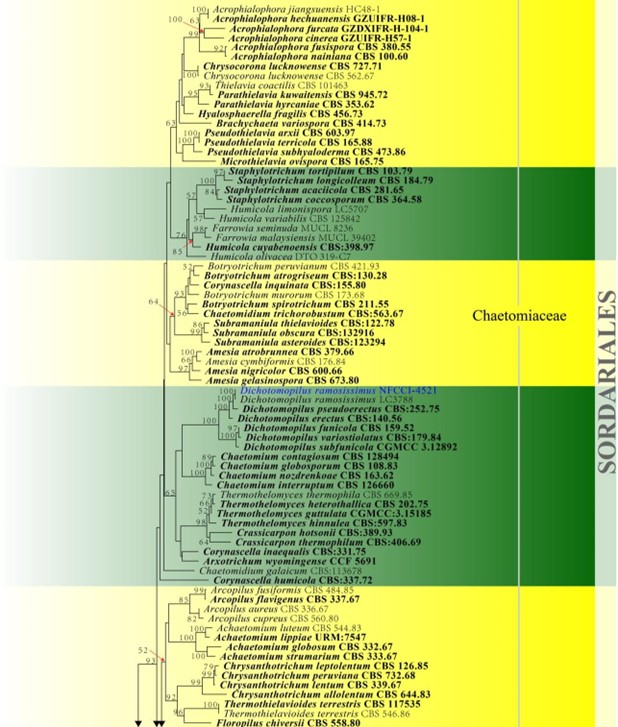
Figure 23 – Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, tub2 and rpb2 sequence data of Sordariales. Related sequences were taken from Wang et al. (2016b). Two hundred and fifty-seven strains are included in the combined analyses which comprised 2717 characters (855 characters for LSU, 480 characters for ITS, 860 characters for tub2, 522 characters for rpb2) after alignment. Members of Amphisphaeriales are used as outgroup
taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. The best RAxML tree with a final likelihood value of -59082.079074 is presented. Estimated base frequencies were as follows: A = 0.238795, C = 0.266829, G = 0.275292, T = 0.219085; substitution rates AC = 1.483472, AG = 3.436969, AT = 1.859704, CG = 1.064423, CT = 6.934931, GT = 1.000000; gamma distribution shape parameter a = 0.757314. Bootstrap support
values for ML greater than 50% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
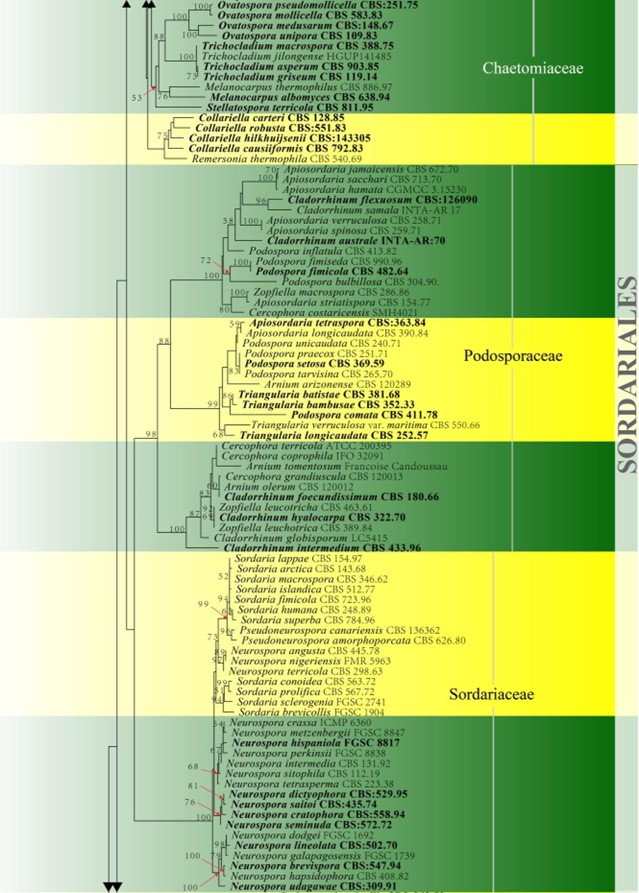
Figure 23 – Continued.
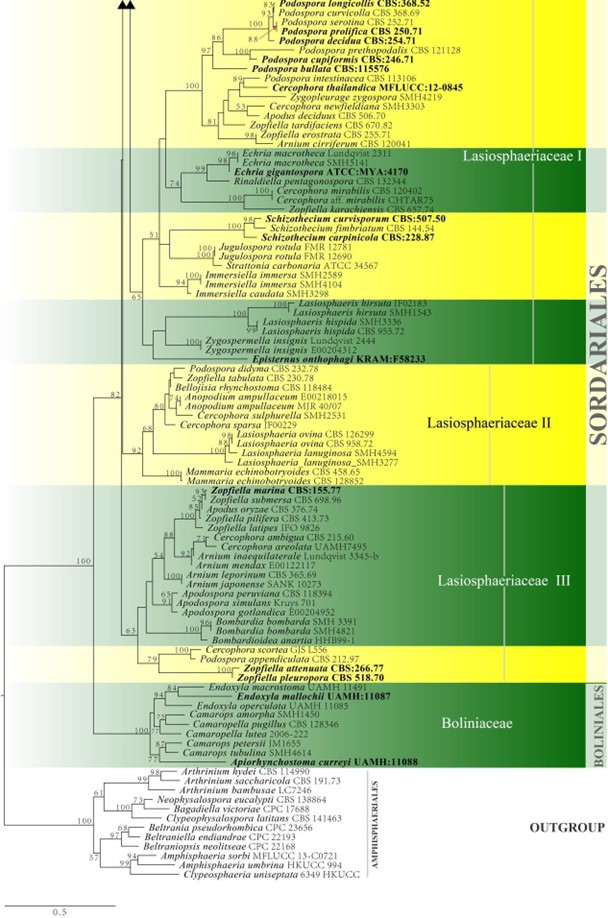
Figure 23 – Continued.
Genera
