Pararamichloridiales Crous, Persoonia 39: 357 (2017)
MycoBank number: MB 823462 ; Index Fungorum number: IF 823462; Facesoffungi number: FoF 06519;
Pararamichloridiales comprises the monotypic family Pararamichloridiaceae and includes a single genus Pararamichloridium. In our phylogenetic analyses generated from LSU and ITS sequence data, Pararamichloridiales formed a monophyletic clade with high statistical support (100% MLBS/1.00PP), which is the same with Crous et al. (2017a) (Fig. 17). Pararamichloridiales is characterised by branched, subhyaline to brown, septate conidiophores, with polyblastic, terminal and intercalary conidiogenous cells that produce solitary, hyaline, aseptate, clavate to ellipsoid conidia (Crous et al. 2017a). In this study, we accept only one family i.e. Pararamichloridiaceae with one genus i.e. Pararamichloridium. The divergence time for Pararamichloridiales is estimated as 101.5 MYA (Fig. 2), which falls in the range of family status. The status of this order may need revision following further study.
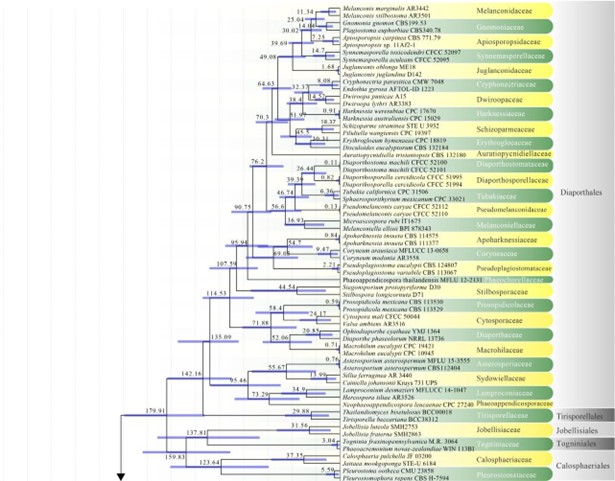
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
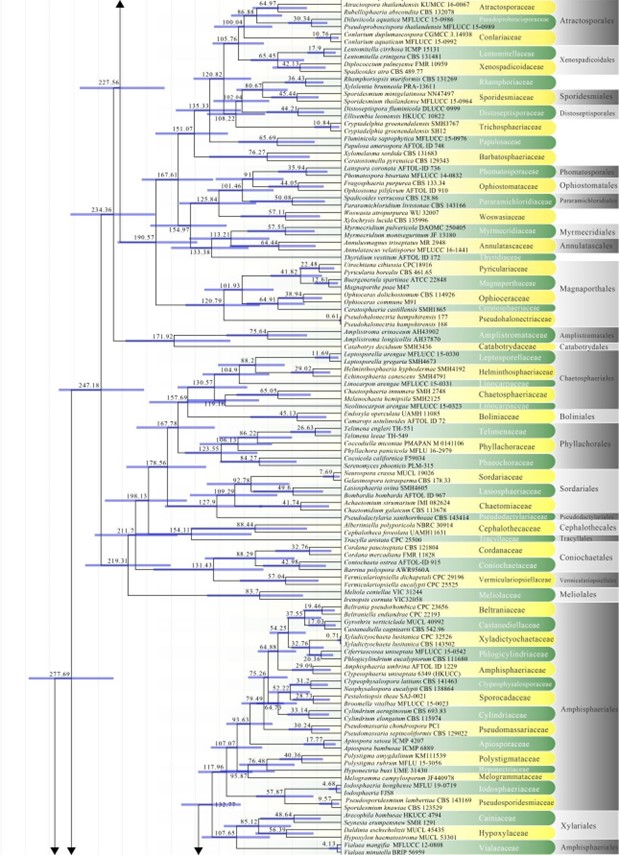
Figure 2 – Continued.
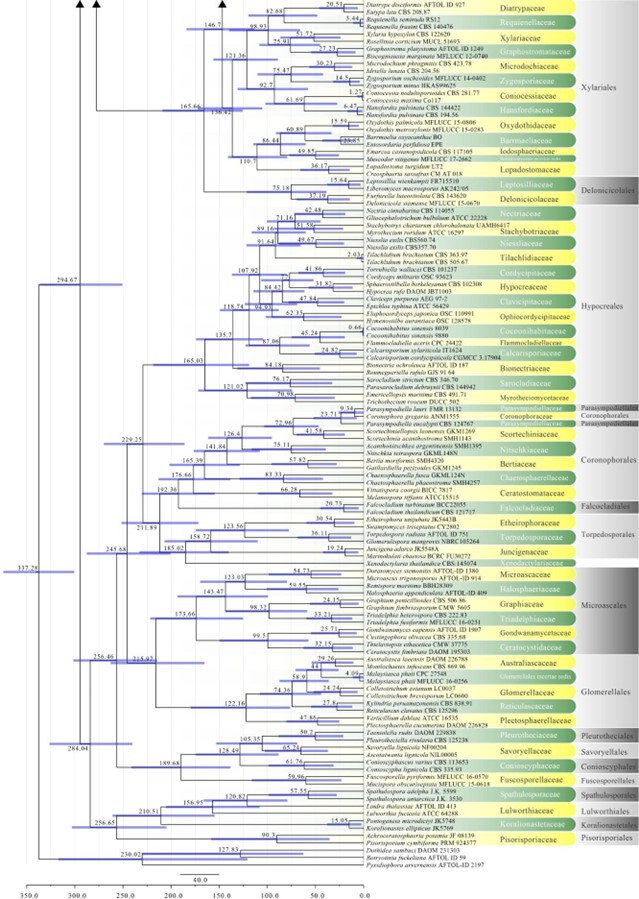
Figure 2 – Continued.
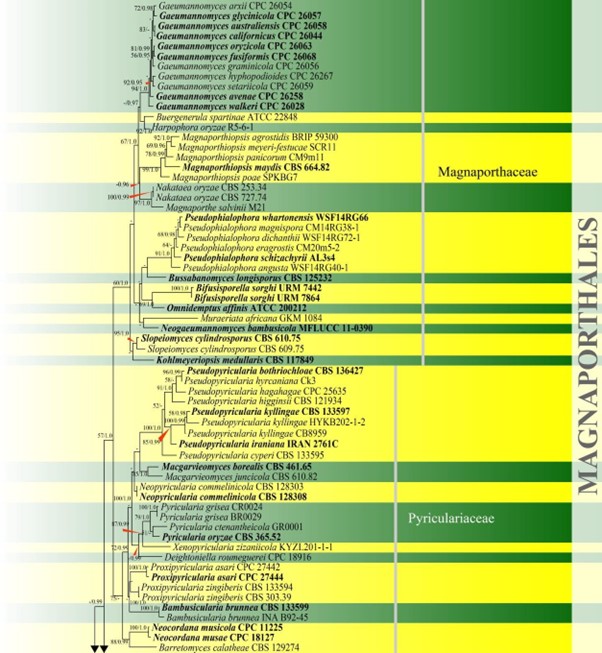
Figure 17 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Magnaporthales and Pararamichloridiales. Eighty-seven strains are included in the combined analyses which comprised 1559 characters (875 characters for LSU, 684 characters for ITS) after alignment. Diaporthe obtusifoliae (CPC 32336) and D. passiflorae (CBS 132527) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 12664.4150 is presented. Estimated base frequencies were as follows: A = 0.2243, C = 0.2819, G = 0.2966, T = 0.1971; substitution rates AC = 1.5257, AG = 2.2337, AT = 2.6637, CG = 1.0862, CT = 7.4361, GT = 1.0000; gamma distribution shape parameter a = 0.4747. Bootstrap support values for ML greater than 50% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
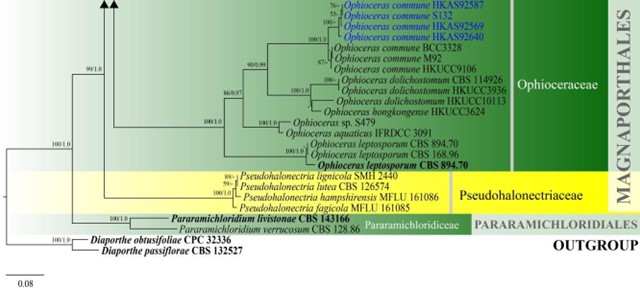
Figure 17 – Continued.
Families
