Paraleptospora chromolaenae Mapook & K.D. Hyde, sp. nov.
MycoBank number: MB 557348; Index Fungorum number: IF 557348; Facesoffungi number: FoF 07811; Fig. 63
Etymology: Name reflects the host genus Chromolaena, from which this species was isolated.
Holotype: MFLU 20-0343
Saprobic on dead stems of Chromolaena odorata. Sexual morph: Ascomata 220–305 µm high × 210–340 µm diam. (x̄ = 265 × 260 µm, n = 5), immersed to semi-immersed, solitary or scattered, gregarious, globose to subglobose, coriaceous, dark brown, appearing as dark spot with red area on host surface. Ostiole short papillate. Peridium 15–20 µm wide, several layers, comprising dense, thick-walled, reddish brown to dark brown, pseudoparenchymatous cells, arranged in textura angularis. Hamathecium comprising 1–2 µm wide, broadly filiform, septate, branching, pseudoparaphyses, anastomosing above the asci. Asci 80–120 × 7.5–11 µm ( x̄ = 100 × 9 µm, n = 30), 8-spored, bitunicate, fissitunicate, cylindrical to cylindric-subclavate, slightly curved, pedicellate, apically rounded, with an ocular chamber. Ascospores 30–40 × 3–4 µm (x̄ = 35 × 3.5 µm, n = 20), overlapping, 1–2-seriate, hyaline to pale yellow, cylindric-fusiform, tapering towards narrow the rounded ends, 6–7-septate, broader at the center and slightly constricted at septa, straight to slightly curved, guttulate, with- out polar appendages. Asexual morph: Undetermined.
Pre-screening for antimicrobial activity: Paraleptospora chromolaenae (MFLUCC 17-1481) showed antimicrobial activity against E. coli with an 8 mm inhibition zone, when compared to the positive control (9 mm), but no inhibition of B. subtilis and M. plumbeus.
Culture characteristics: Ascospores germinating on MEA within 48 h. at room temperature and germ tubes produced from both ends. Colonies on MEA circular, mycelium umbonate, cultures white at the surface, creamy-white in reverse (Fig. 65a).
Material examined: THAILAND, Lampang Province, Ngao, on dead stems of Chromolaena odorata, 21 September 2016, A. Mapook (LP3, MFLU 20-0343, holotype); ex- type culture MFLUCC 17-1481.
GenBank numbers: LSU: MN994563, ITS: MN994586, SSU: MN994609, TEF1: MN998167
Notes: In a BLASTn search of NCBI GenBank, the closest match with the ITS sequence of Paraleptospora chromolaenae (MFLUCC 17-1481, ex-holotype) with 91.36% similarity was Phaeosphaeriaceae sp. (strain MUT 4404, KC339239). The closest match with the LSU sequence with 99.34% similarity was Neostagonospora arrhenatheri (strain MFLUCC 15–0464, KX910091). The closest match with the SSU sequence with 99.71% similarity was Parastagonospora nodorum (strain LSNZN10, MH269310), while the closest match with the TEF1 sequence with 94.91% similarity was Yunnanensis phragmitis (strain MFLUCC 17-0365, MF683625). In the present phylogenetic analysis, P. chromolaenae clusters with P. chromolaenicola with high bootstrap support (100% ML and 1.00 BYPP, Fig. 52). However, P. chromolaenae differs from P. chromolaenicola in having slightly larger ascomata (220–305 × 210–340 µm vs. 140–310 × 120–300 µm) and slightly larger ascospores (30–40 × 3–4 µm vs. 28–38 × 3–4 µm) with 6–7-septa, and without polar appendages, while P. chromolaenicola has (5–)7–8-septate ascospores, and very few ascospores with polar appendages (Table 12). A comparison of the ITS (+5.8S) gene region of P. chromolaenae and P. chromolaenicola reveals 23 base pair differences (4.6%) across 503 nucleotides. Therefore, P. chromolaenae is described as a new species based on phylogeny and morphological comparison.
Table 12 Synopsis of Paraleptospora species with similar morphological features discussed in this study
| Species | Asocomata (µm) | Peridium (µm) | Asci (µm) | Ascospores (µm) | Septation of ascospores | References |
| P. chromo-laenae (MFLUCC 17-1481)
|
220–305 high × 210–340 diam. | 15–20 | 80–120 × 7.5–11 | 30–40 × 3–4 | 6–7-septate | This study |
| P. chromo- laenicola (MFLUCC 17-1450)
|
140–310 high × 120–300 diam | 14–27.5 | 100–120 × 8–11 | 28–38 × 3–4 | (5–)7–8-septate | This study |
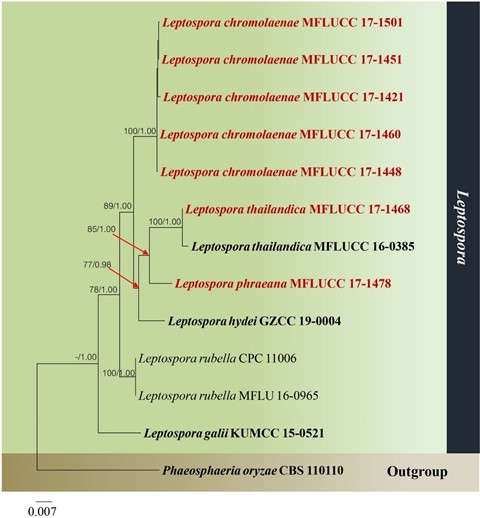
Fig. 52 Phylogram generated from maximum likelihood analysis based on combined dataset of LSU, ITS, SSU, TEF1 and RPB2 sequence data. Thirteen strains are included in the combined sequence analysis, which comprise 4416 characters with gaps. Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of − 8333.195072 is presented. The matrix had 327 distinct alignment pat- terns, with 24.82% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.252190, C = 0.233838, G = 0.264447, T = 0.249525; substitution rates: AC = 1.127465, AG = 2.895211, AT = 2.022869, CG = 0.534488, CT = 8.448227, GT = 1.000000; gamma distribution shape parameter α = 0.020000. Boot- strap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes. Newly generated sequences are in dark red bold and type species are in bold. Phaeosphaeria oryzae (CBS 110110) is used as outgroup taxon
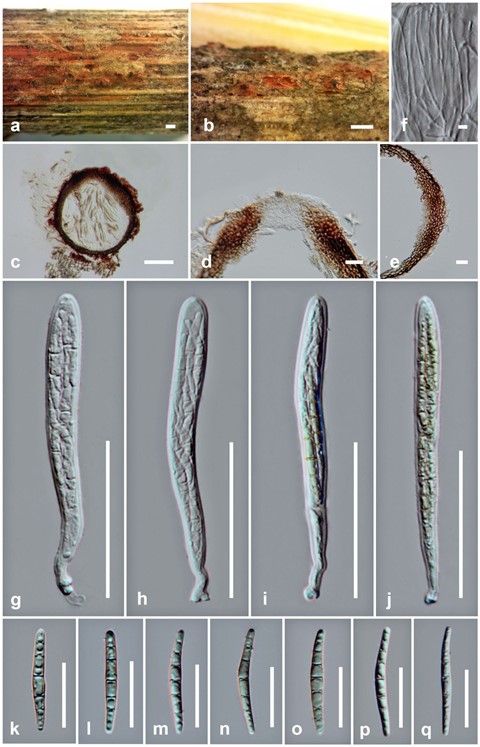
Fig. 63 Paraleptospora chromolaenae (holotype) a, b Appearance of ascomata on substrate. c Section through ascoma. d Ostiole. e Peridium. f Pseudoparaphyses. g–j Asci. k–q Ascospores. Scale bars: a, b = 500 µm, c = 100 µm, g–j = 50 µm, d, e, k–q = 20 µm, f = 5 µm
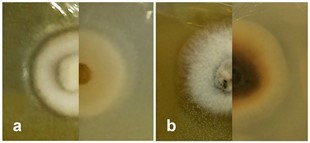
Fig. 65 Culture characteristics on MEA: a Paraleptospora chromolaenae (MFLUCC 17-1481), b Paraleptospora chromolaenicola (MFLUCC 17-1450)
