Parafusicladium amoenum (R.F. Casta~neda & Dugan) Crous, M. Shen & Y. Zhang ter, comb. nov.
MycoBank number: MB 831544; Index Fungorum number: IF 831544; Facesoffungi number: FoF 12044; Fig. 7.
Basionym: Anungitopsis amoena R.F. Castaneda & Dugan, Mycotaxon 72: 118. 1999.
Synonyms: Fusicladium amoenum (R.F. Casta~neda & Dugan) Crous et al., Stud. Mycol. 58: 207. 2007. Cladosporium amoenum R.F. Casta~neda, BCCM MUCL Agroindustrial fungi-yeasts. 1998. Nom. inval., Art. 38.1(a) (Shenzhen).
Description and illustration: Untereiner et al. (1998), Ho et al. (1999), Crous et al. (2007b).
Typus: Cuba, Santiago de Cuba, La Gran Piedra, fallen leaves of Eucalyptus sp. (Myrtaceae), 2 Nov. 1994, R.F. Casta~neda (Ho et al. 1999: 117, Figs 2, 3, holotype; epitype ATCC 200947 (designated in Ho et al. 1999), culture ex-epitype CBS 254.95 = ATCC 200947 = IMI 367525 = INIFAT C94/155 = MUCL 39143).
Notes: Cladosporium amoenum was first described from fallen leaves of Eucalyptus sp. collected in Cuba, which was, unfortunately, invalid because it lacked a Latin diagnosis (Untereiner et al. 1998). Ho et al. (1999) validated its name and assigned it to Anungitopsis (as A. amoena), which was subsequently assigned to Fusicladium (as F. amoenum) (Crous et al. 2007b). The colony of Fusicladium amoenum is pseudocladosporium-like, while the loci of the conidiogenous cells are neither prominently thickened, nor refractive (Ho et al. 1999, Crous et al. 2007a). Phylogenetically, Fusicladium amoenum clusters in Parafusicladium, sister to P. paraamoenum (Fig. 1).

Fig 1. Consensus phylogram (50 % majority rule) of 691 952 trees resulting from a Bayesian analysis of the combined alignment of ITS, LSU, tef1, tub2 and rpb2 sequences of Venturiales. Bayesian posterior probabilities (PP) > 0.80 are shown at the nodes and the scale bar represents the expected changes per site. Some branches were shortened to facilitate layout. The tree was rooted with Microthyrium microscopicum (CBS 115976). Culture collection numbers, substrates and countries are indicated behind the species names. Those highlighted in bold are new taxa or new combinations proposed in this study, and type strains are marked with “T” (ex-type in black, ex-epitype in red). Relevant morphological characteristics plotted are abbreviated as follows: Sol – conidia solitary, C – conidia in chains, NS – aseptate conidia, 1S – 1-septate conidia, MS – multi- septate conidia (septa ≥ 2), St – straight or slightly curved conidia, Coil – coiled conidia, Y – Y-shaped conidia; Ramo – ramoconidia present, NR – ramoconidia not observed; ? – asexual morphology not available (either from references or from sporulation induced in this study); and morphological characters plotted in red means strains failed to sporulate in this study and plotted values are taken from the original description, observation of this study or related references. Other characteristics are explained in the legend.
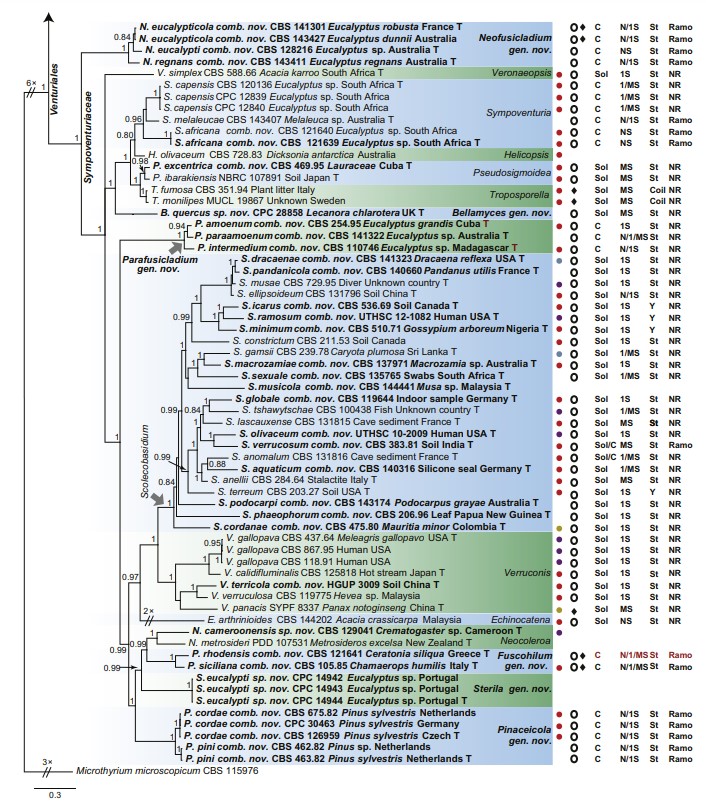
Fig 1. (Continued).
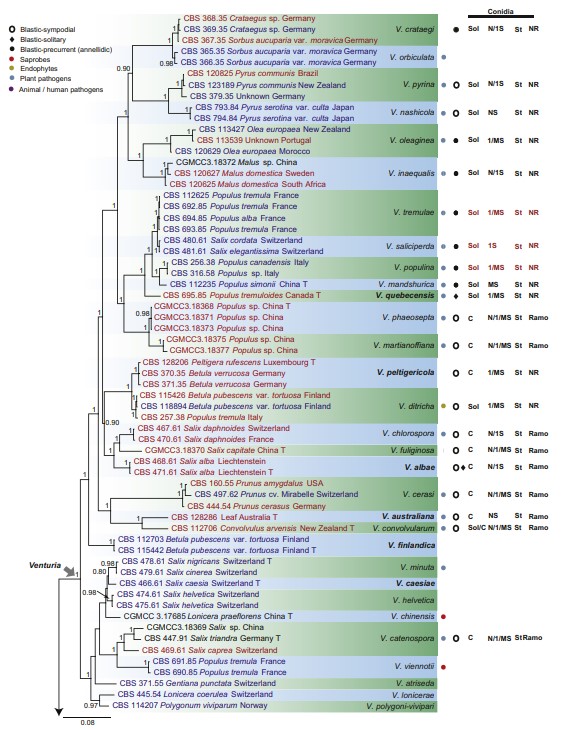
Fig 2. Consensus phylogram (50 % majority rule) of 42 902 trees resulting from a Bayesian analysis of the combined alignment of ITS, LSU, tef1, tub2 and rpb2 sequences of Venturiaceae. Bayesian posterior probabilities (PP) > 0.80 are shown at the nodes and the scale bar represents the expected changes per site. Some branches were shortened to facilitate layout. The tree was rooted with Pseudoanungitea vaccinii (CBS 143164). See title of Fig. 1 for an explanation of the characters plotted on the tree. Strains in red text sporulated in this study, while those in blue text failed to sporulate and those in black text were not studied.
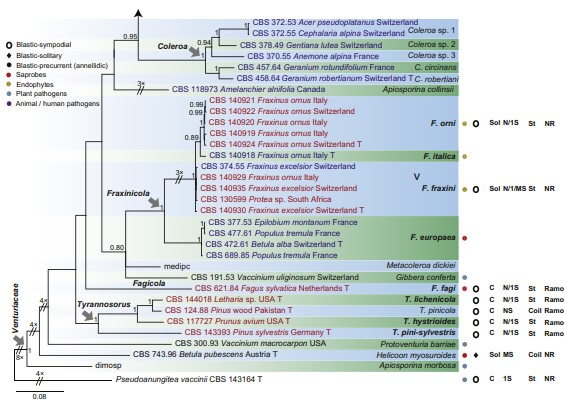
Fig 2. (Continued).
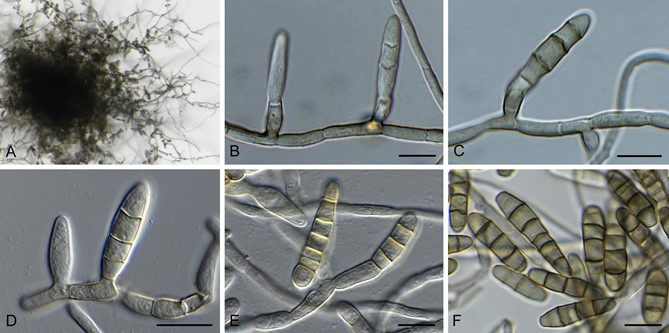
Fig. 3. Bellamyces quercus (culture ex-type CPC 28858) asexual morph. A. Colony on OA. B–E. Conidiogenous cells producing conidia. F. Multi-septate conidia. Scale bars: B–F = 10 μm.
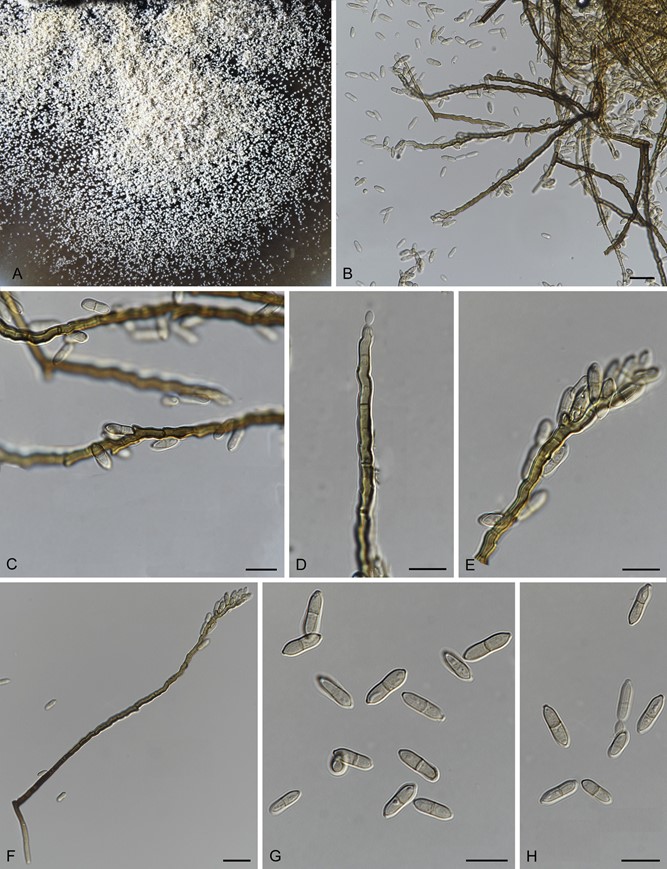
Fig. 7. Parafusicladium amoenum (culture ex-type CBS 254.95) asexual morph. A. Colony on OA. B–F. Long conidiophores reduced to sympodial conidiogenous cells. G, H. Pale brown and 1-septate conidia. Scale bars: B– H = 10 μm.
