Neodictyosporium macarangae Tennakoon, C.H. Kuo & K.D. Hyde, , in Tennakoon, Kuo, Maharachchikumbura, Thambugala, Gentekaki, Phillips, Bhat, Wanasinghe, de Silva, Promputtha & Hyde, Fungal Diversity 108: 1–215 (2021)
MycoBank number: MB 555755; Index Fungorum number: IF 555755; Facesoffungi number: FoF 09378; Fig. 1
Etymology: Name reflects the host genus Macaranga.
Holotype: MFLU 18-0086
Saprobic on dead leaf petiole of Macaranga tanarius. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial, on the upper surface of the petiole 95–120 × 230–270 μm (x̄ = 108 × 250 μm, n = 10), solitary, scattered, superficial, globose to subglobose, black. Peridium thin at the apex, base and sides 9–12 μm (x̄ = 10.5 μm, n = 8), a single layer, composed of brown to black thin-walled cells of textura angularis. Conidiogenous cells (2–) 3–4.5 μm broad (x̄ = 3.5 μm, n = 10), holoblastic, integrated, smooth, brown, producing a branched conidium at the tip, cup-shaped or doliiform. Conidia two types, Type I: dictyosporous, 22–26 (–32) × (11–) 13–17 μm ( x̄ = 24 × 15.5 μm, n = 10), light brown, regularly consisting of 5–6 rows of cells, each rows comprising 6 cells, 3.1–3.6 μm wide; Type II: clavate to obovoid, acrogenous, 22–30 (–32) × 16–22 μm (x̄ = 27 × 18 μm, n = 10), hyaline to sub-hyaline, 3-septate, guttulate, smooth-walled.
Culture characteristics: Colonies on PDA, 20–25 mm diam. after 3 weeks, colonies from above: medium dense, irregular, effuse, surface slightly rough with undulate margin, smooth, dark brown to black at the margin, light brown to cream at the centre; reverse dark brown to black at the margin, light brown to black at the centre; mycelium light brown to black with tufting.
Material examined: Taiwan, Chiayi, Ali Shan Mountain, Fanlu Township area, Dahu forest, dead leaf petiole of Macaranga tanarius (Euphorbiaceae), 05 August 2019, S. Tennakoon, R007 (MFLU 18–0086, holotype); ex- type living culture, MFLUCC 17–2653, ibid., 15 August 2019, R007B (NCYU 19-0355, paratype); ex-paratype living culture, NCYUCC 19-0232, ibid., 17 August 2019, R007C (NCYU 19-0362, paratype); living culture, NCY- UCC 19-0339.
GenBank numbers: MFLUCC 17–2653: LSU: MW114442; SSU: MW114308. NCYUCC 19-0232: LSU: MW114443; SSU: MW114309. NCYUCC 19-0339: LSU: MW114444; SSU: MW114310.
Notes: The morphology of Neodictyosporium resembles Dictyosporium in having cheirosporous conidia (hand-like) (Goh 1999; Boonmee et al. 2016). Asexual morph is hyphomycetous and conidia form directly on mycelia or substrate, whereas Neodictyosporium conidia form in pycnidia. Two types of conidia can be seen in Neodictyosporium species, such as cheirosporous conidia (type I) and clavate to obovoid conidia (type II) (Fig. 143). Phylogeny (LSU, SSU, RPB2) indicates that our collection (MFLUCC 17–2653, NCYUCC 19-0232, NCYUCC 19-0339 and NCYUCC 19-0340) nested within Sordariomycetes, between Acrodictys and Platytrachelon, all of which form a clade with 80% ML, 0.95 BYPP statistical support (Fig. 2). Neodictyosporium can be distinguished from Acrodictys and Platytrachelon, in having two types of conidia (cheirosporous conidia and clavate to obovoid conidia), whereas Acrodictys has broadly clavate, obovoid to pyriform, muriform conidia (Luo et al. 2019) and Platytrachelon has cylindrical, one-celled, strongly curved conidia (Réblová 2013).

Fig. 1 Neodictyosporium macarangae (MFLU 18-0086, holotype): a, b Conidiomata on host. c Vertical section of a conidioma. d Peridium. e, f Conidiogenous cells and developing conidia. g Squash mount of conidia. h–j Conidia (Type I and II). k–m Conidia (Type II). n A germinating conidium. o Colonies from above (on PDA). p Colonies from below (on PDA). Scale bars: c = 50 μm, d = 8 μm, e, f = 10 μm, g = 30 μm, h–n = 10 μm
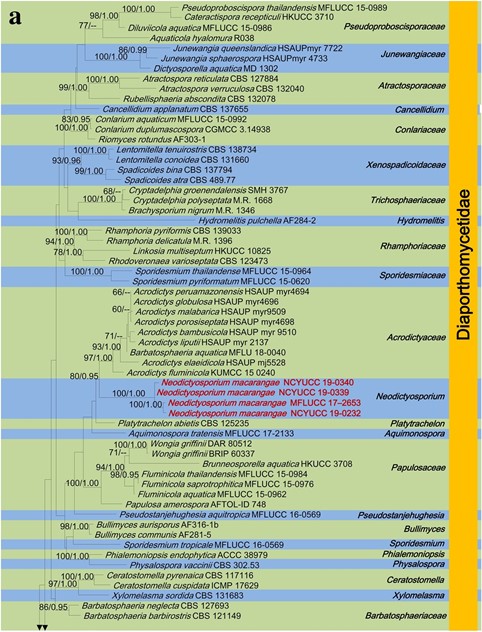
Fig. 2 The best scoring RAxML tree with a final likelihood value of – 53787.710415 for combined dataset of LSU, SSU and RPB2 sequence data. The topology and clade stability of the combined gene analyses was compared to the single gene analyses. The tree is rooted Leotia lubrica (AFTOL-ID 1), Microglossum rufum (AFTOL- ID 1292). The matrix had 1928 distinct alignment patterns with 42.93% undetermined characters and gaps. Estimated base frequencies were as follows; A= 0.253562, C = 0.234832, G = 0.288461, T = 0.223146; substitution rates AC = 1.308095, AG = 2.828270, AT = 1.119351, CG = 1.218357, CT= 7.175485, GT = 1.000000;gamma distribution shape parameter α = 0.567661. Newly generated sequences are in red. Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.90 are given above the nodes
