Neoacladium indicum P.N. Singh & S.K. Singh, sp. nov.
MycoBank number: MB 556211; Index Fungorum number: IF 556211; Facesoffungi number: FoF 5688; Figs. 139, 141
Etymology: Specific epithet ‘‘indicum’’ refers to the country of origin.
Holotype: AMH 10054
Saprobic on dead bark of Leucaena leucocephala L. Asexual morph Colonies uniformly spread on dead bark, yellow, velvety. Conidiophores semi-macronematous, mononematous, fasciculate, dichotomously branched, loosely interwoven, straight to flexuous, subhyaline to light olivaceous, smooth-walled, septate, 337–752 9 9–13 lm. clavate, obclavate, pyriform, lenticular, sometimes broadly ellipsoidal to ampulliform, pigmented, smooth-walled, base truncate, subhyaline to light olivaceous, 10.5– 25.88 9 9.36–16.5 lm (x¯ = 15.75 9 13.55 lm, n = 30).Sexual morph Undetermined
Culture characteristics: Conidia germinating on malt extract agar (MEA). Colonies light yellow (4A4), reaching 4.2 cm diam. in 10 days at 25 °C, rhizoidal with irregular margin, surface powdery, reverse greyish orange (5B5). Hyphae septate, unbranched to branched, pigmented, smooth and thin walled, subhyaline to light olivaceous, 5–5.62 lm wide.
Material examined: INDIA, Maharashtra, Pune District, on dead bark of Leucaena leucocephala (Fabaceae), 30 July 2017, P.N. Singh; (AMH 10054, holotype), ex-type living culture (NFCCI 4480).
GenBank numbers: ITS: MK391496, LSU: MK391493.
Notes: The proposed taxon is compared with the asexual genus Acladium in having subhyaline to light olivaceous ampulliform to pyriform conidia produced from tubular to inflated and conidiogenus cells which are persistent, and occasionally sessile. Chlamydospores in the proposed taxon produced laterally and form trident like structure, which are pigmented. Most of these features are absent in Acladium. However, the conidial shape, length–width are very much variable in N. indicum, which separate it from other reported taxa in this group. Proposed taxon is also distinct from other genera in this group, like Burgoa, Minimedusa, Ceratobasidium, (Rogers 1935; Weresub and LeClair 1971) in having non-bulbuliferous vegetative parenchymatous propagules. During this study the sexual stage Botryobasidium of Acladium was not encountered.
The proposed N. indicum (AMH 10054 holotype), is different from other Botryobasidium species based on the sequence analyses. On megablast analysis, ITS sequence of N. indicum showed 80% (524/651) identity and 53 gaps (8%) with B. subcoronatum, 84% (343/410) identity and 27 gaps (6%) with B. simile (GEL2348), and 81% (521/645) identity 54 gaps (8%) with B. intertextum UC2022959. The phylogenetic analysis clearly establishes Neoacladium as a novel genus, and N. indicum as the type species with strong bootstrap values (Figs. 142, 143).
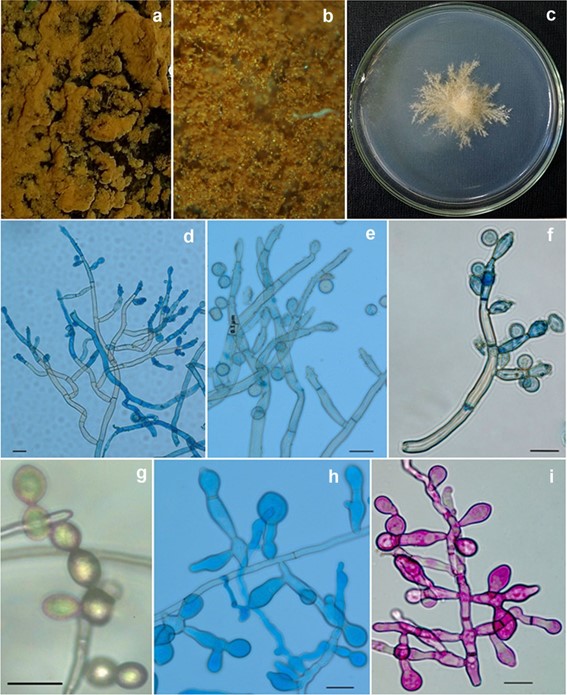
Fig. 139 Neoacladium indicum (AMH 10054, holotype). a Yellow velvety fungal colonies spreaded on outer bark surface. b Stereoscopic surface view of fungal colonies from substratum. c Colony morphol- ogy (above view). d Dichotomously branched conidiophores. e En- larged microscopic view of dichotomously branched conidiophores with intercalary, lateral and terminal dentate conidiogenous cells and conidia. f A detached branch of conidiophore bearing conidiogenous cells and conidia. g Catenate conidia. h–i Trident shaped and inflated chlamydospores. Scale bars: d–i = 20 lm
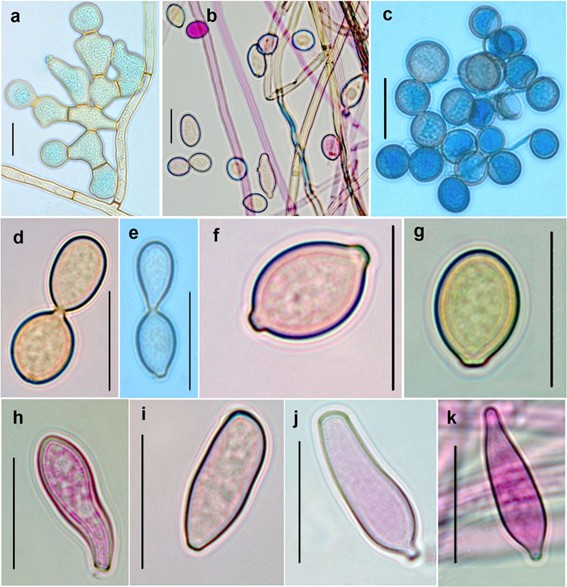
Fig. 141 Neoacladium indicum (AMH 10054, holotype). a Branched chlamydospores. b Hyphae and different types of conidia. c Numerous globose to sub-globose conidia. d, e Catenate conidia. f Lenticular conidium. g Pyriform conidium. h, i Clavate conidia. j, k Obclavate conidia with protuberant hila. Scale bars: a–k = 20 lm
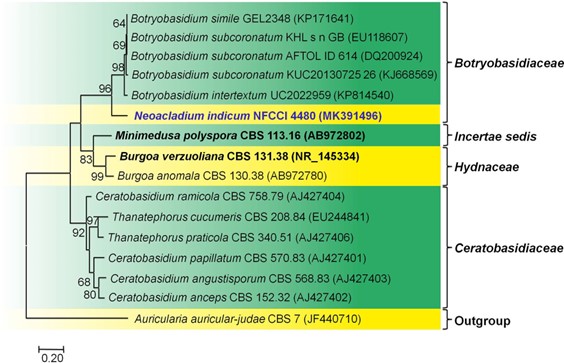
Fig. 142 Molecular phylogenetic analysis of ITS gene region of Neoacladium indicum (AMH 10054, holotype) by Maximum likeli- hood method. Sixteen strains are included in the sequence analyses which comprise 668 positions after alignment. Auricularia auricular– judae CBS 7 (JF440710) (Auriculariaceae, Auriculariales) is used as the outgroup taxon. Bootstrap values for maximum likelihood (ML) greater than 50 are placed along the branches, respectively. The evolutionary history was inferred by using the Maximum likelihood method based on the Jukes-Cantor model (Jukes and Cantor 1969). The tree with the highest log likelihood (- 4900.5990) is shown. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). The ex-type strains are in bold and black. The newly generated sequence is indicated in bold and blue
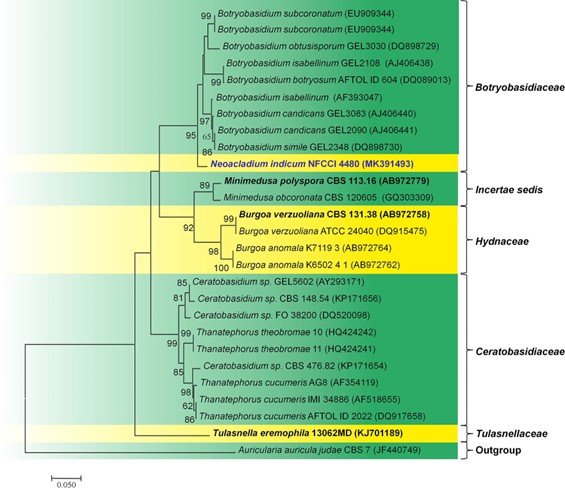
Fig. 143 Molecular Phylogenetic analysis of LSU of Neoacladium indicum (AMH 10054, holotype) by Maximum likelihood method. Twenty seven strains are included in the sequence analyses which comprise 2176 positions after alignment. Auricularia auricular–judae CBS 7 (JF440710) (Auriculariaceae, Auriculariales) is used as the outgroup taxon. Bootstrap values for maximum likelihood (ML) greater than 50 are placed along the branches respectively. The evolutionary history was inferred by using the Maximum likelihood method based on the Kimura 2-parameter model (Kimura 1980). The tree with the highest log likelihood (- 3161.4740) is shown. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). The ex-type strains are in bold and black. The newly generated sequence is indicated in bold and blue
Species
