Meliolales Gäum. ex D. Hawksw. & O.E. Erikss., Syst. Ascom. 5(1): 180 (1986)
Index Fungorum number: IF 90483; MycoBank number: MB 90483; Facesoffungi number: FoF 09682;
Meliolales was introduced by Hawksworth & Eriksson (1986), accommodating a single family Meliolaceae. Hosagoudar (2003) introduced Armatellaceae as a new family in this order based on morphology. Lumbsch & Huhndorf (2010) placed Meliolales in the class Sordariomycetes. This was confirmed and followed by Hongsanan et al. (2015, 2017) and Maharachchikumbura et al. (2016b). Hongsanan et al. (2015) provided a recent monograph that reappraised the genera of Meliolales. Zeng et al. (2017) provided a checklist for identifying Meliolales species, including the current names of host plants with their corresponding Meliolales species. Currently, Meliolales comprises two families, viz. Armatellaceae and Meliolaceae, but sequence data is only available for Meliolaceae. The phylogenetic relationship of Meliolaceae in the current study is different from Hongsanan et al. (2015) and Zeng et al. (2018) with regards to the position of Irenopsis, which is sister to the main Meliola clade (Fig. 19). All recent studies indicated that Meliola is polyphyletic and the clade including some Meliola species clusters in the Appendiculella and Asteridiella lineages with low bootstrap support. The divergence time for Meliolales is estimated as 219 MYA (Fig. 2). Currently, there are two families and nine genera in this order (this paper).
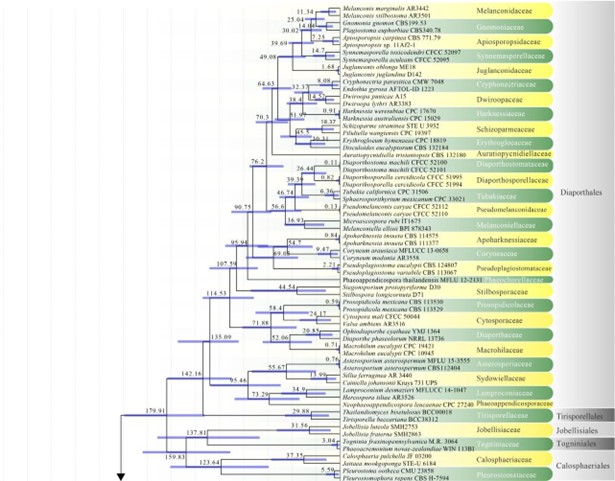
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
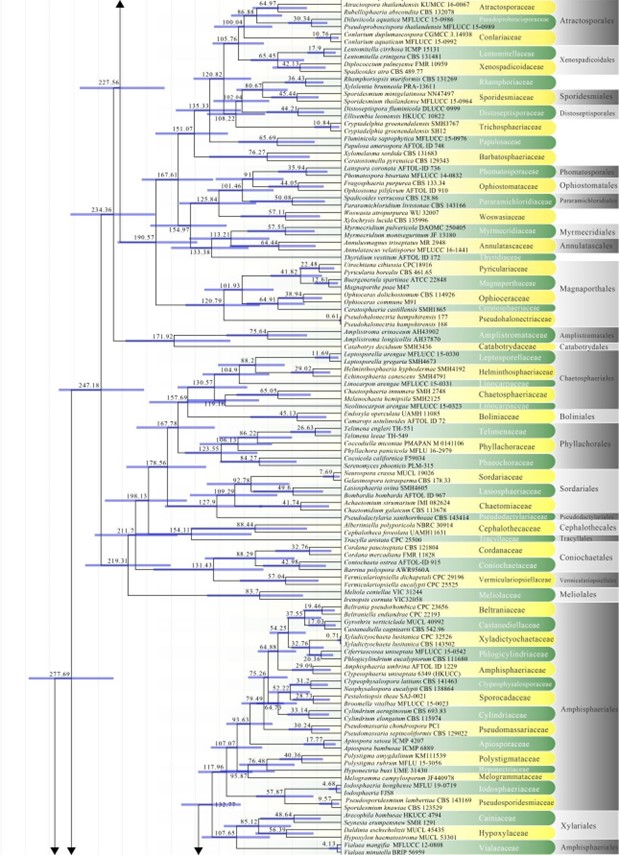
Figure 2 – Continued.
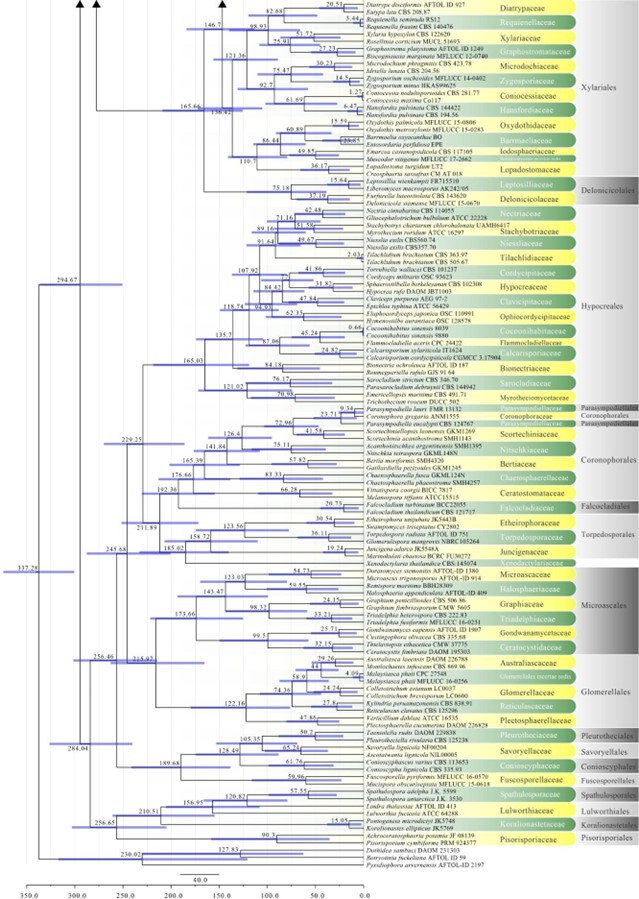
Figure 2 – Continued.
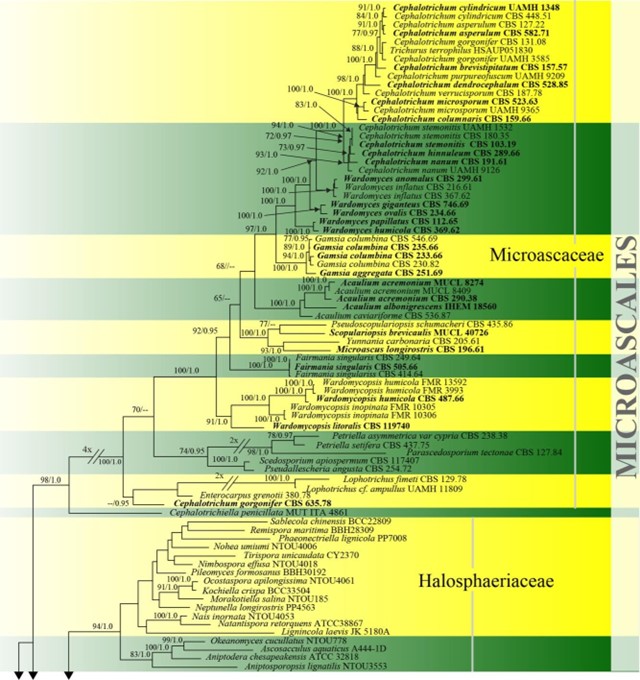
Figure 19 – Phylogram generated from maximum likelihood analysis based on combined ITS, LSU, SSU and tef1 sequence data of Microascales. One hundred and thirty-two strains are included in the combined analyses which comprised 4538 characters (653 characters for ITS, 1215 characters for LSU, 1685 characters for SSU, 985 characters for tef1) after alignment. Lecanicillium fungicola var. aleophilum (CBS 357.80) and L. fungicola var. fungicola (CBS 992.69) (Cordycipitaceae, Hypocreales) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of -46610.749560 is presented. Estimated base frequencies were as follows: A = 0.240650, C = 0.256729, G = 0.270623, T = 0.231998; substitution rates AC = 0.965367, AG = 2.062871, AT = 1.418856, CG = 0.960832, CT = 4.902766, GT = 1.000000; gamma distribution shape parameter a = 0.478984. Bootstrap support values for ML greater than 65% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold.
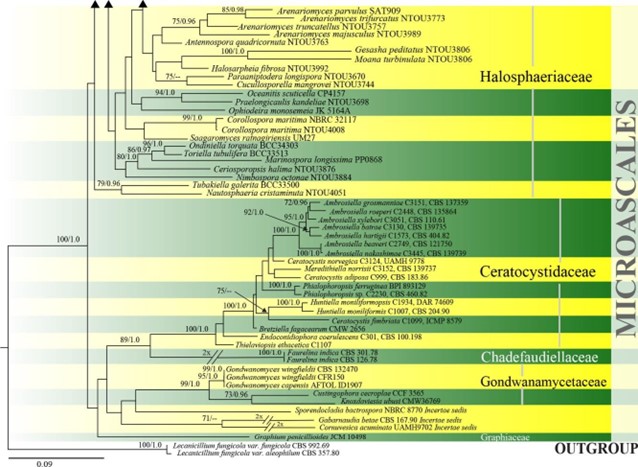
Figure 19 – Continued.
Families
