Lulworthiales Kohlm., Spatafora & Volkm.-Kohlm., Mycologia 92(3): 456 (2000)
MycoBank number: MB 90552; Index Fungorum number: IF 90552; Facesoffungi number: FoF 09684;
Lulworthiales was introduced based on morphology and LSU and SSU phylogeny to accommodate the genera Lulworthia and Lindra, which were initially accommodated in Halosphaeriales (Kohlmeyer et al. 2000). All taxa referred to this order are marine aquatic fungi. The order is accommodated in Lulworthiomycetidae, where it forms a strongly-supported clade with Koralionastetales (Maharachchikumbura et al. 2016b) with a divergent age of 289 MYA (Hongsanan et al. 2017). Lulworthia is polyphyletic, as observed from the combined LSU, SSU and ITS phylogeny (Fig. 16). This is in accordance with other studies which also reported the polyphyly of Lulworthia (Jones et al. 2008, 2009, 2019, Abdel-Wahab et al. 2010, Azevedo et al. 2017). Spathulosporaceae, typified by Spathulospora is also reported to group in Lulworthiales (Jones et al. 2019). Spathulosporaceae was initially placed in Spathulosporales based on morphology (Kohlmeyer 1973), but molecular data of some Spathulosporaceae species have shown that the taxa have a higher affinity to Lulworthiales, even though the type species of Spathulosporaceae was not included, since it lacks sequence data (Inderbitzin et al. 2004, Campbell et al. 2005, Jones et al. 2009). Currently there is one family (the placement of Spathulosporaceae is not confirmed in Lulworthiales) and 15 genera in this order with both asexual and sexual morphs (this paper). Further studies are required to resolve the higher order rank of the marine algicolous parasites Spathulospora.
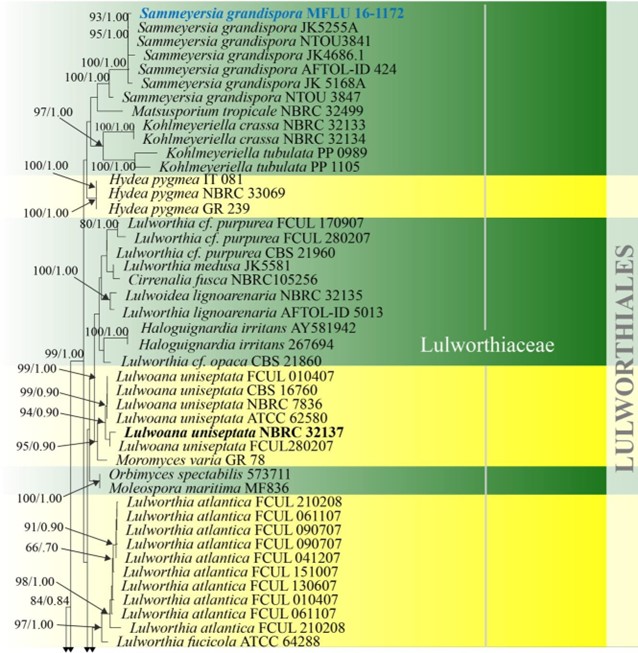
Figure 16 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU and ITS sequence data of selected taxa from Koralionastetales, Lulworthiales and Pisorisporiales. Sixty-nine strains are included in the combined gene analyses comprising 2780 characters after alignment (930 characters for LSU, 1059 characters for SSU and 784 characters for ITS). Fuscosporella pyriformis (MFLUCC 16-0570) and Parafuscosporella mucosa (MFLUCC 16- 0571) are used as outgroup taxa. Analyses of each single gene were performed and the topology of each tree had clade stability. The tree topology of the maximum likelihood was similar to the Bayesian and maximum parsimony analyses. Maximum likelihood analysis with 1000 bootstrap replicates yielded a best tree with the likelihood value of -24852.531353. The matrix had 1582 distinct alignment patterns, with 38.88% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.249996, C = 0.239207, G = 0.291483, T = 0.219314; substitution rates AC = 1.016404, AG = 2.006035, AT = 1.090073, CG = 1.243791, CT = 5.331398, GT = 1.000000; gamma distribution shape parameter α = 0.395174. Maximum parsimony (black) and maximum likelihood (black) bootstrap values >65% and Bayesian posterior probabilities (green) >0.90 (ML/BYPP) are given above the nodes. Ex-type strains are in bold and new strains are in blue.
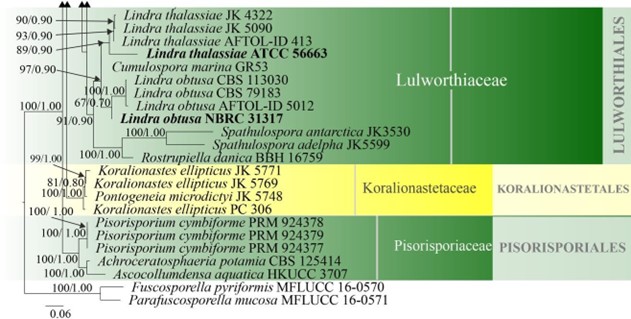
Figure 16 – Continued.
Famillies
