Lopadostoma (Nitschke) Traverso, Fl. ital. crypt. 1(2): 169 (1906)
Index Fungorum number: IF 2925; MycoBank number: MB 2925; Facesoffungi number: FoF 00706; 24 morphological species (Species Fungorum 2020), 11 species with sequence data.
Type species – Lopadostoma turgidum (Pers.) Traverso
Notes – Lopadostoma comprises 12 species from Europe which are saprobes producing pseudostromatic sexual morphs and libertella-like asexual morphs (Fig. 152, Jaklitsch et al. 2014). So far species delimitation of Lopadostoma is based only on ITS and LSU sequence data with low resolution, while protein coding regions, especially rpb2, are essential for further species delimitation (Daranagama et al. 2016, Jaklitsch et al. 2014). The Lopadostoma gastrinum sexual morph and L. turgidum asexual morph are illustrated in this entry (Fig. 151, 152).
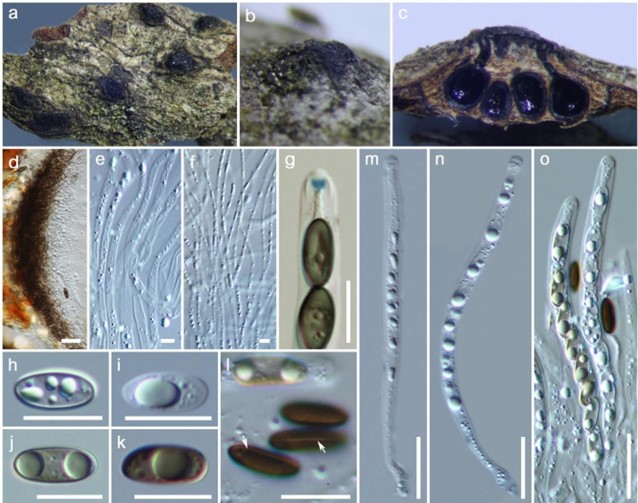
Figure 151 – Lopadostoma gastrinum (Material examined – ITALY, Province of Forlì-Cesena [FC], Monte Mirabello – Predappio, on dead fallen branch of Quercus sp. (Fagaceae), 12 April 2017, Erio Camporesi IT3269C, MFLU 17-0941; ibid. HKAS 102310). a, b Stromata on the host. c Cross section of stroma. d Peridium. e, f Paraphyses. g J+, apical ring in Melzer’s reagent. h-l Ascospores (l-white arrows show germ slit). m-o Asci. Scale bars: d, m-o = 20 μm, g-l = 10 μm, e, f = 5 μm.
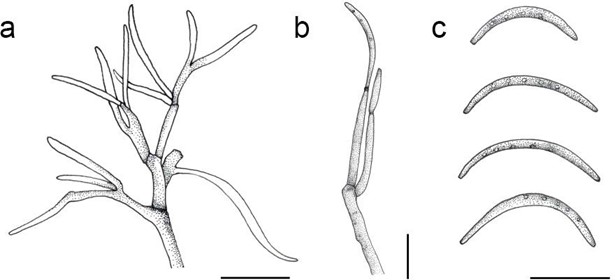
Figure 152 – Lopadostoma turgidum (ex-epitype culture CBS 133207). a, b Conidiophores (b. showing phialide bearing conidium). c Conidia. Scale bars: 10 μm (redrawn from Jaklitsch et al. 2014).
Species
Lopadostoma turgidum
