Leptosporellaceae S. Konta & K.D. Hyde, Mycosphere 8(10): 1956 (2017)
Index Fungorum number: IF 553956; MycoBank number: MB 553956; Facesoffungi number: FoF 03840; 11 species.
Saprobic or endophytic on various host plants. Sexual morph: Ascomata solitary, superficial, comprising black, subglobose, carbonaceous, dome-shaped, raised, blister-like areas within the plant tissues, flattened at the base, ostiole central. Peridium outer cells merge with the host epidermal cells, composed of dark brown to black cells of textura angularis. Paraphyses numerous, hyaline, hypha-like, septate, longer than asci. Asci 8-spored, unitunicate, cylindrical, long-pedicellate, with a J-, wedge-shaped, subapical ring. Ascospores fasciculate, hyaline or pale-yellowish in mass, spiral, filiform, straight or curved, aseptate, ends rounded, with or without polar mucilaginous appendages. Asexual morph: Undetermined (adapted from Konta et al. 2017).
Type genus – Leptosporella Penz. & Sacc.
Notes – The family was introduced by Konta et al. (2017) in Chaetosphaeriales with support from the analysis of combined LSU and ITS sequence data. In this study, a new species, Leptosporella elaeidis, is introduced (Fig. 8).

Figure 8 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Chaetosphaeriales and Tracyllalales taxa. Ninety-six strains are included in the combined analyses which comprised 1695 characters (1081 characters for LSU, 614 characters
for ITS) after alignment. Neurospora crassa MUCL 19026 and Gelasinospora tetrasperma CBS 178.33 (Sordariaceae, Sordariales) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 23777.689886 is presented. Estimated base frequencies were as follows: A = 0.231060, C = 0.264793, G = 0.308265, T = 0.195882; substitution rates AC = 1.388486, AG = 1.836207, AT = 1.649563, CG = 0.971659, CT = 6.316962, GT = 1.000000; gamma distribution shape parameter a = 0.460297. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
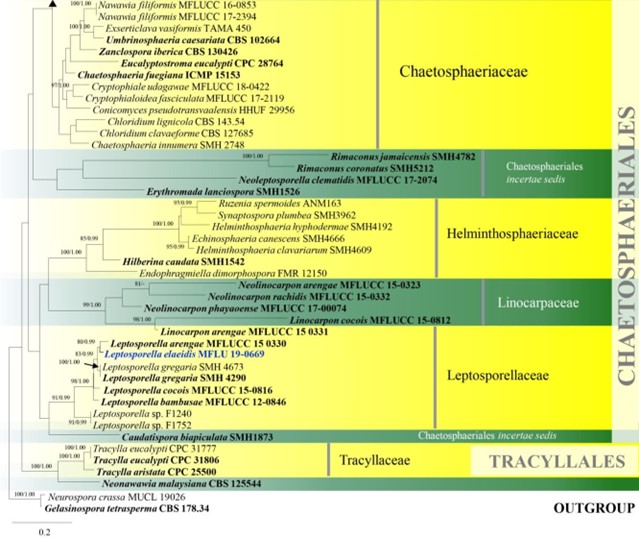
Figure 8 – Continued.
Genera
