Kaarikia C. Mayers & T.C. Harr., gen. nov.
Index Fungorum number: MB 824946; Facesoffungi number: FoF
Typification: Kaarikia abrahamsonii C. Mayers & T. C. Harr.
Etymology: After Aino Käärik (born 1918 Aino Mathiesen; also known as Aino Mathiesen-Käärik), for her work with the fungi associated with many forest insects including the discovery of P. ferruginea from T. lineatum, and in recognition of her recently cele- brated 100th birthday.
Solitary, obovoid, truncate, sometimes septate coni- dia formed terminally on simple conidiophores. Thick- walled chlamydospores, terminal or intercalary, globose to irregular, inside-pigmented hyphae. Sensitive to cycloheximide. Sexual state unknown.
Notes: – Kaarikia is unique in morphology and DNA sequences, necessitating treatment as a monotypic genus. Phylogenetic analyses place Kaarikia within the Sordariomycetes with uncertain placement in order and family (FIG. 9), although it has phylogenetic affinity with the aquatic Distoseptispora (Distoseptisporaceae) (Su et al. 2016; Luo et al. 2018; Yang et al. 2018) and Aquapteridospora lignicola (Yang et al. 2015) as well as Taeniolella sabalicola (Delgado and Miller 2017). It is also related to Ellisembia leonensis, which has been informally treated in Distoseptispora (Su et al. 2016; Lueo et al. 2018; Yang et al. 2018) but never formally combined. Kaarikia resembles Distoseptispora species in general culture morphology and in having thick, darkly pigmented hyphae and multiseptate conidia, but Kaarikia does not produce Sporidesmium-like conidio- phores; its conidiophores are significantly less devel- oped, its conidia much less pigmented, and its formation of spherical chlamydospores unique. The production of chlamydospores in Kaarikia does strongly resemble the conidiogeny of some lichenicolous Taeniolella (Heuchert et al. 2018), for which DNA sequences are not available. However, species of this paraphyletic genus are not known to be related to the Distoseptisporaceae (Ertz et al. 2016), except for T. sabalicola, which is embedded in Distoseptispora (FIG. 9) but does not share the morphology of the lichenicolous taxa (Delgado and Miller 2017) nor belong in Taeniolella sensu stricto (Heuchert et al. 2018). Because the inclusion of neighboring taxa in the family Distoseptisporaceae is ambiguous, and Kaarikia does not match the morphological description of the family, we treat Kaarikia as genus incertae sedis.
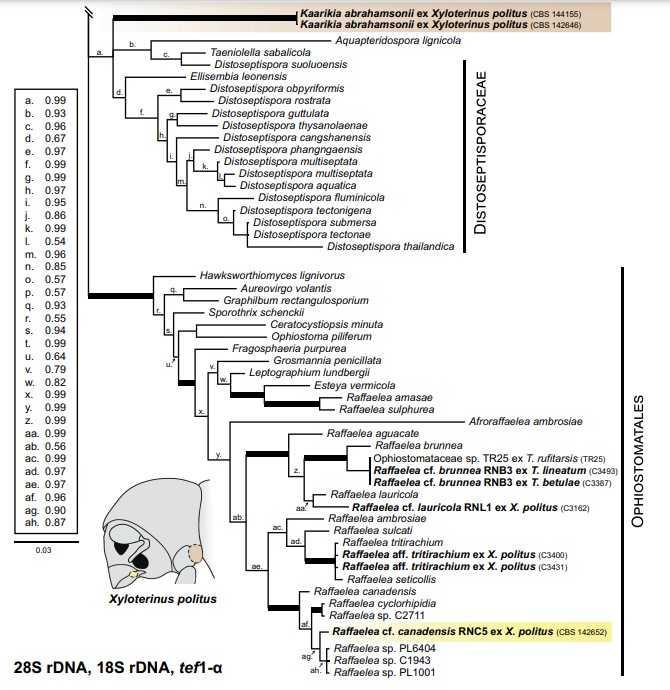
Figure 9. Excerpted multilocus tree from Bayesian analysis of the combined 28S rDNA, 18S rDNA, and tef1-α data set of the two mycangial mutualists of Xyloterinus politus (Kaarikia abrahamsonii and Raffaelea cf. canadensis RNC5) and other representative members of the Diaporthomycetidae. The two mycangial mutualists, and other Raffaelea isolated from Xyloterini in this study, are in bold. The mycangial mutualists are in shaded boxes. A generic Xyloterini silhouette shows the general positions of the two types of mycangia in X. politus, and the mycangia are shaded the same color as the box of their respective mutualists. Posterior probabilities from Bayesian analysis are indicated in the key and correspond to lowercase letters on branches; values are rounded to two significant digits, except posterior probability values ≥0.995 and <1.0, which are represented as “0.99”. Thickened branches indicate Bayesian posterior probability of 1.0 and are not included in the key. Families and orders are in small caps and indicated with vertical bars. For bolded species, accession numbers in the Westerdijk Fungal Biodiversity Institute (CBS), US National Fungus Collections (BPI), or Iowa State University collection (C) are provided; collection information for other species representatives are detailed in SUPPLEMENTARY TABLE 2. For Raffaelea cf. brunnea RNB3, only the sequence of isolate C3387 was used as a representative sequence in analysis, but two other isolates (C3493 and TR25) with identical (100% identity) 28S sequences are represented in the figure. The full tree, which is rooted to Dothidea sambuci, is available in SUPPLEMENTARY FIG. 2. Bar = 0.03 estimated substitutions per site.
Species
