Haplohelminthosporium Konta & K.D. Hyde, gen. nov
Index Fungorum number: IF557873; Facesoffungi number: FoF09169
Etymology:—Haplo in Greek means single, which refers to the single conidium in each conidiophore. It is a close relative of Helminthosporium.
Saprobic on living leaves and petioles of Calamus sp. On living leaves, small spots, circular to irregular, yellow in the beginning, later becoming red brown surrounded by yellow. Colonies on natural substrate forming black patches on the upper leaf, petiole surfaces.
Sexual morph:Undetermined.
Asexual morph:Hyphomycetous.
Colonies on natural substrate forming black patches on the upper leaf, petiole surfaces. Mycelium mostly immersed, partly on the surface forming small stroma-like aggregations of red brown pseudoparenchymatous cells. Conidiophores arising singly or fasciculate from stroma cells, erect, simple, unbranched, straight, curved and swollen at apex, septate, thick-walled, cylindrical, smooth, bulbous at base, hyaline in the middle, brown to yellow-brown at 1–2-cells above the base, pale brown to yellow-brown at apical cell. Conidiogenous cells monotretic, terminal, determinate, cylindrical, wide and yellow-brown with a well-defined, small, noncicatrized pore at the apex. Conidia one for each conidiophore, obpyriform to lageniform, straight or curved, smooth, olive-brown, distoseptate, with a dark scar at the base.
Type species:—Haplohelminthosporium calami Konta & K.D. Hyde
Notes: – Haplohelminthosporium is established as a monotypic genus with Hap. calami as the type species. ITS phylogenetic analyses separated this genus from other genera, while in the LSU and multigene analyses it clustered with Helminthosporium and Helminthosporiella, but both without good statistical support (Figures 1 and 2). Haplohelminthosporium is presented herein as an asexual morph (hyphomycete) similar to Helminthosporium and Helminthosporiella in that it is hyphomycete with an erect conidiophore, monotretic conidio- genous cell and distoseptate conidia [19,22,63]. The type species of Helminthosporium has pale to dark brown, septate conidiophores, with terminal and intercalary polytretic conidio- genous cells, noncicatrized pores at the apex and upper 3–4 cells, solitary or short catenate conidia that are subhyaline to brown, distoseptate, and is dark brown to black scar at the base [19]. Helminthosporiella has brown to red-brown conidiophores with terminal, poly- tretic conidiogenous cells, with catenate and easily disarticulating chains of conidia that are medium brown, striated at surface and distoseptate [63]. However, Haplohelminthosporium is distinguished by its unbranched conidiophores arising solitarily or fasciculate from the stroma-like bulbous basal cells that are hyaline in the middle, brown to red-brown at 1–2-cells above the base, pale brown to red-brown and curved at the apical cell with well-defined non-cicatrized small pores and with a single olive-brown conidium arising from each conidiophore (Figure 3). In the BLAST search of GenBank, the closest match of the LSU, ITS, and SSU sequence data were identical to Helminthosporium spp. Based on distinguishing morphological characteristics together with single/multigene phylogenetic analyses we introduce the newly described strain as a new genus Haplohelminthosporium in Massarinaceae.
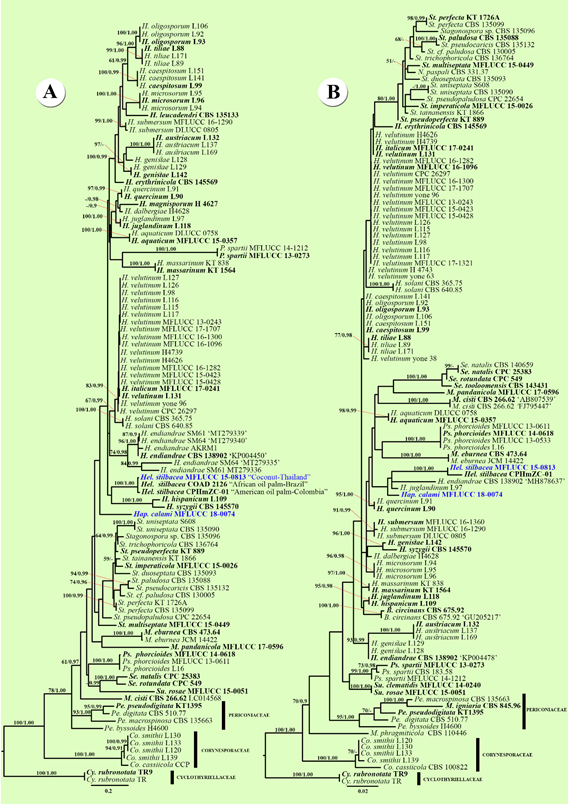
Figure 1. Comparison of the topology of Maximum likelihood majority rule consensus tree for the analyses of some selected Corynesporaceae, Massarinaceae, and Perioconiaceae isolates. (A) Phylogenetic tree of the dataset for ITS sequence data.
(B) Phylogenetic tree of the dataset for LSU sequence data. Bootstrap support values for maximum likelihood (ML) equal to or higher than 50%, and Bayesian Posterior Probabilities (BYPP) equal to or greater than 0.90 are given above each branch. Novel taxa are in blue. Ex-type strains are in bold. The tree is rooted to Cyclothyriella rubronotata strains TR, TR9 (Cyclothyriellaceae).
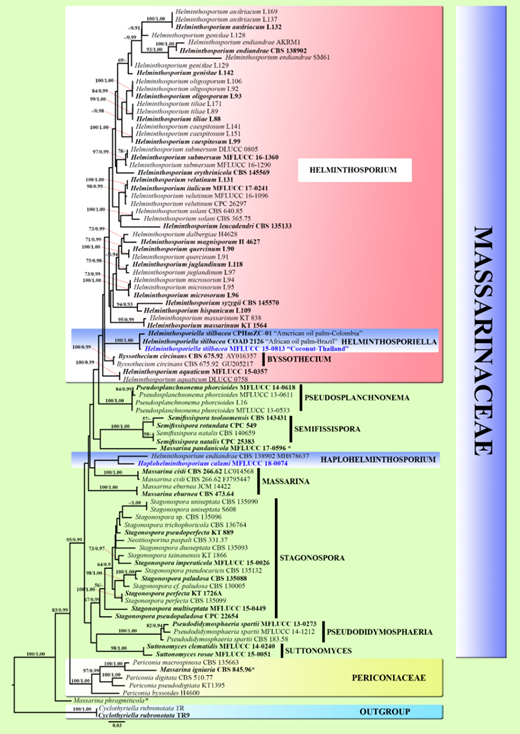
Figure 2. Maximum likelihood majority rule consensus tree for the analyses of Massarinaceae and sister family Perioco- niaceae isolates based on a dataset of combined ITS, LSU, SSU, and tef1-α sequence data. Bootstrap support values for maximum likelihood (ML) equal to or higher than 50%, and Bayesian posterior probabilities (BYPP) equal to or greater than 0.90 are given above each branch. Novel taxa are in blue. Ex-type strains are in bold. The tree is rooted to Cyclothyriella rubronotata strains TR, TR9 (Cyclothyriellaceae).

Figure 3. Haplohelminthosporium calami (MFLU 20-0520, holotype) (A) The forest in Krabi Province. (B–E) Fresh and herbarium palm samples. (F,G) Colonies on living leaf. (H–L) Conidiophores. (M–U) Conidia. (V,W) Germinated conidia.
(X) Culture on PDA. (Y) Conidiophore and conidia on culture. (Z) Conidiogenesis. (AA) Conidiophores. (AB,AC) Conidia. Scale bars: C, E =2 cm, H–W, Y–AC = 50 µm.
Species
