Elongaticonidia W.J. Li, E. Camporesi & K.D. Hyde, gen. nov.
Index Fungorum number: IF557147, Facesoffungi number: FoF 07359
Etymology: Referring to the conidial shape is elongated. Saprobic on dead spines of Rosa canina.
Sexual morph: undetermined.
Asexual morph: Conidiomata black, pyc- nidial, solitary to gregarious, subepidermal, immersed, globose to subglobose, ostiolate. Ostiole short, centrally located. Conidiomatal wall composed of thick-walled, dark brown to brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells form- ing from inner layers of conidiomata, pale brown to hyaline, forming from inner wall layers of conidiomata, pale brown to hyaline, holothallic, ampulliform to cylindrical, discrete, determinate, smooth-walled, consisting of basal thicker- walled cells which become flared at the inception of conid- iogenesis. Conidia hyaline, arthric, formed by disarticulation of the conidial chain, produced in simple unbranched chains with the youngest conidium at the base, elongated, 1-septate, deeply constricted at septum, bearing unbranced, cellular, flexuous, appendages.
Type species: Elongaticonidia rosae W.J. Li, E Campo- resi & K.D. Hyde, sp. nov.
Notes: – Phylogenetic study based on LSU sequence data shows Elongaticonidia close to Mulderomyces (Fig. 7), but it formed a seprate branch in Ostropales (Lecanoro- mycetes). Morpholgically, Elongaticonidia is distinct from Mulderomyces in form of conidia and conidiogenous cells.
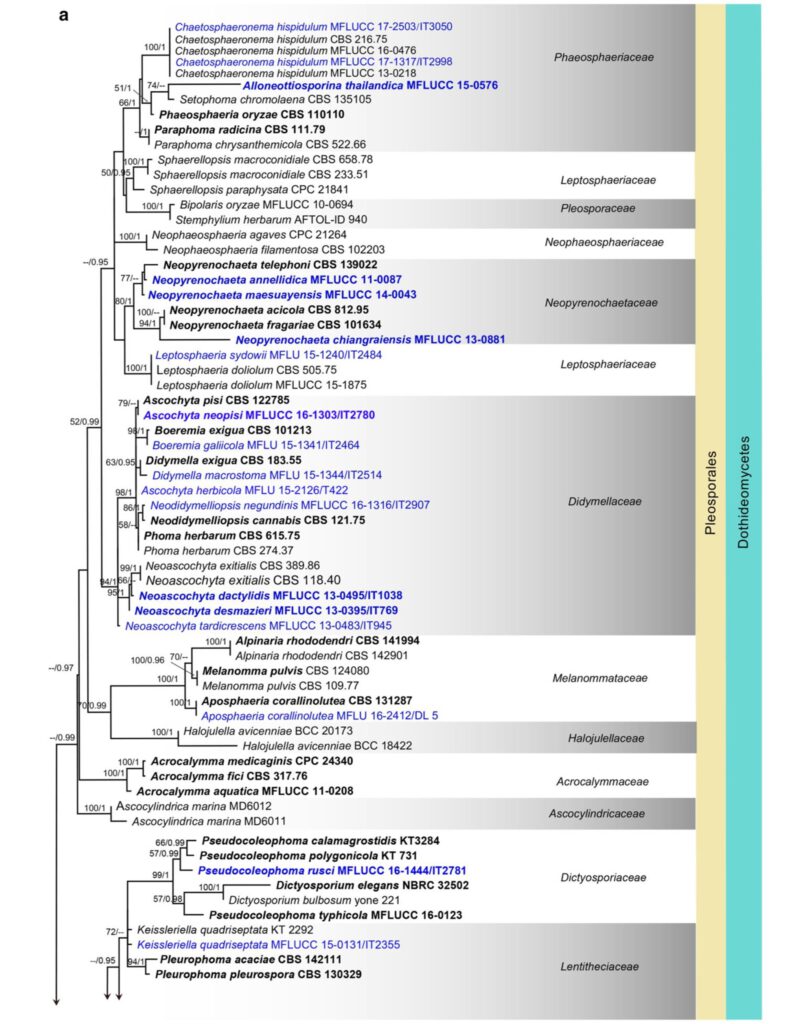
Fig. 7 Phylogenetic tree generated from a maximum likelihood analysis based on LSU sequences data of Agaricomycetes (Basidi- omycota), Coniocybomycetes, Dothideomycetes, Eurotiomycetes, Geoglossomycetes, Lecanoromycetes, Leotiomycetes, Lichinomy-cetes, Orbiliomycetes, Pezizomycetes and Sordariomycetes (Ascomy-cota). Related sequences were obtained from GenBank. Six hundred and twenty-nine strains are included in the analyses, which comprise 978 characters including gaps. Phytophthora moyootj VHS27218 is used as the outgroup taxon. The tree topology of the maximum likeli-hood analysis is similar to the maximum parsimony and the Bayesian analysis. The best scoring RAxML tree with a fnal optimization likelihood value of −2919.850785 is presented. The matrix had 245 distinct alignment patterns, with 14.17% of undetermined characters or gaps. Estimated base frequencies were: as follows: A=0.209309,C=0.308850, G=0.253017, T=0.228825; substitution rates AC=0.826247, AG=2.476259, AT=1.076270, CG=0.884896, CT=4.252047, GT=1.000000; gamma distribution shape parameter α=0.168455. Maximum likelihood (MLBS) bootstrap support values higher than 50%, and Bayesian posterior probabilities ≥ 0.95 (PP) are shown above or below the nodes. Hyphen (“–”) indicates a value lower than 50% for MPBS and MLBS and a posterior probability lower than 0.95 for BYY. The scale bar indicates 10 changes. Ex-type or ex-epitype strains are in bold. New isolates are in blue
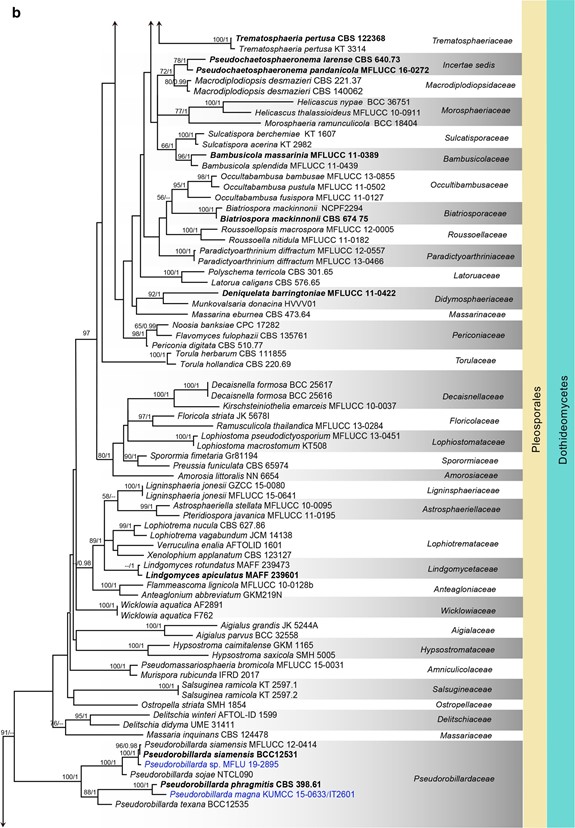
Fig. 7 (continued)
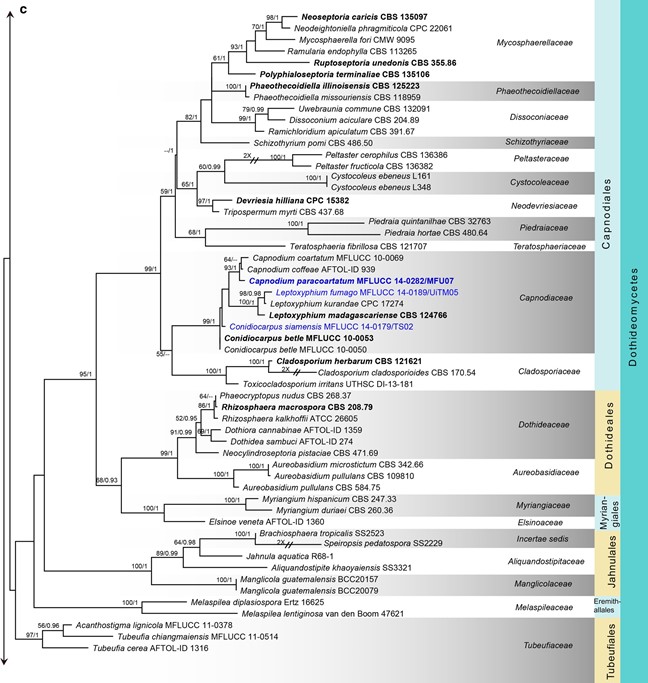
Fig. 7 (continued)
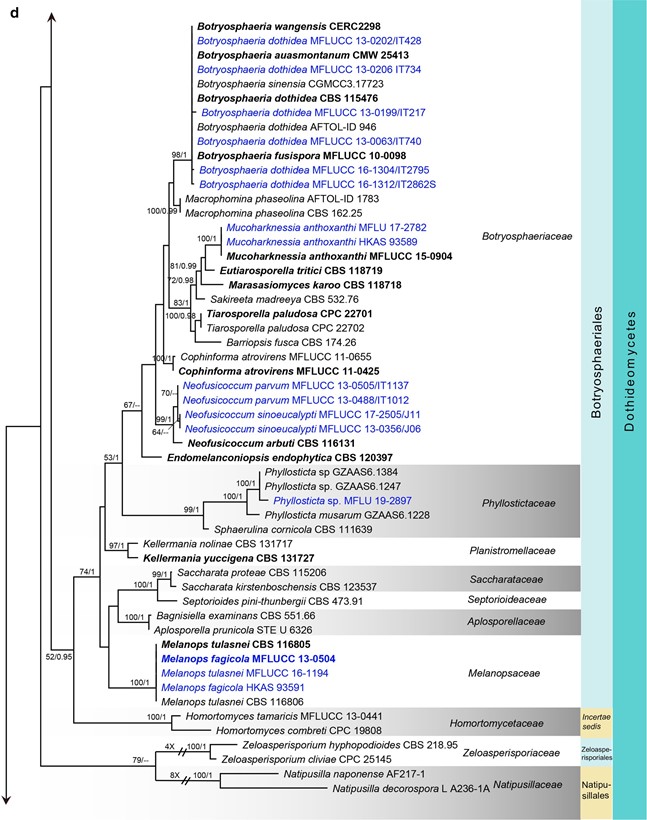
Fig. 7 (continued)
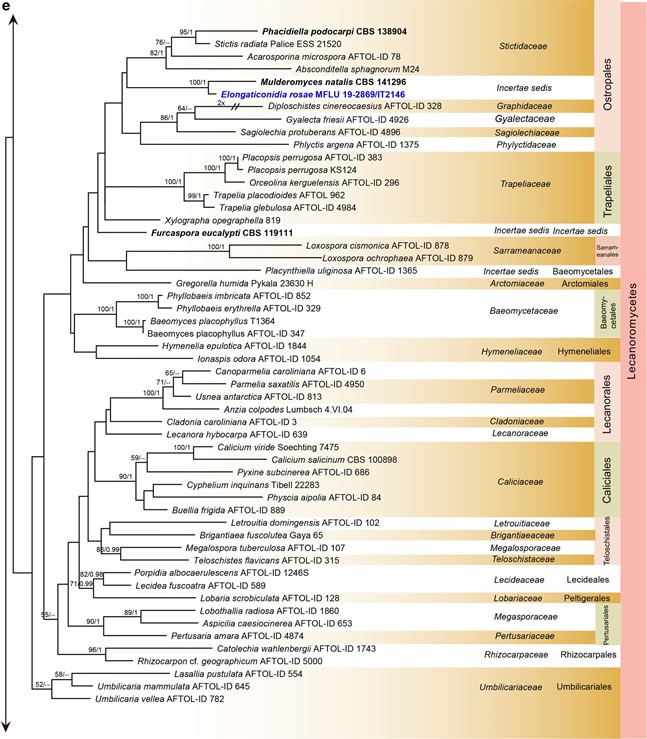
Fig. 7 (continued)
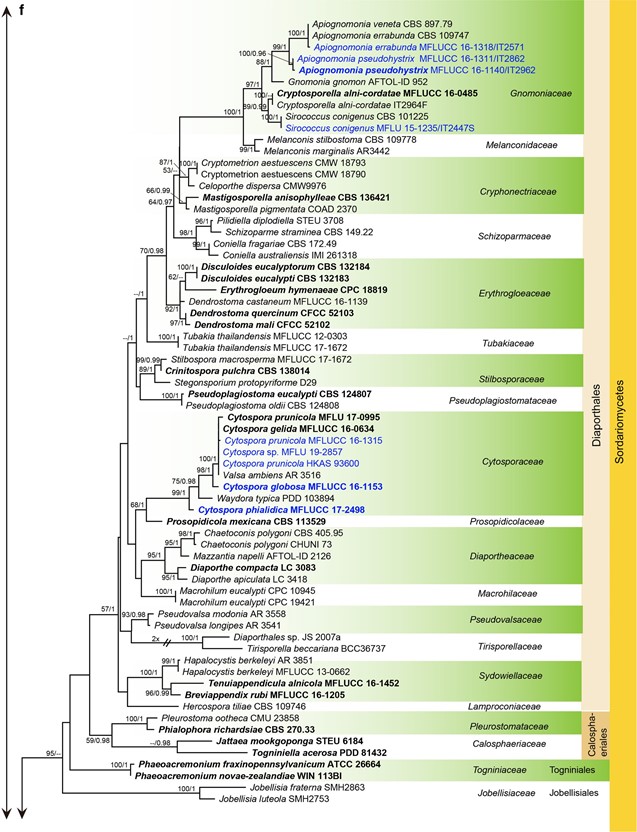
Fig. 7 (continued)
Fig. 7 (continued)
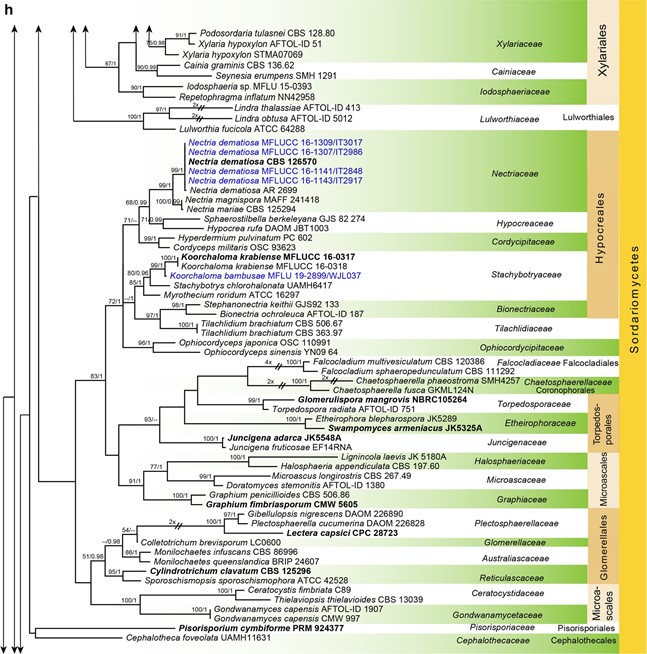
Fig. 7 (continued)
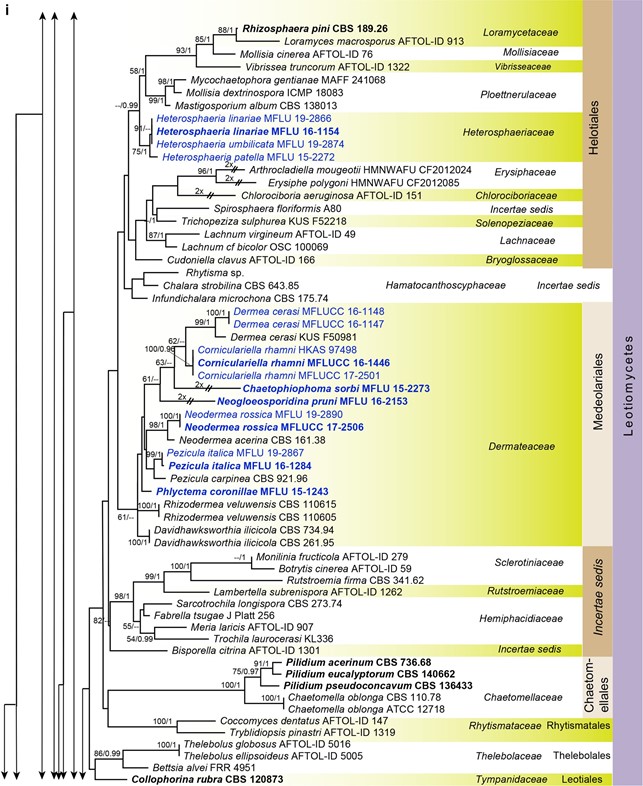
Fig. 7 (continued)
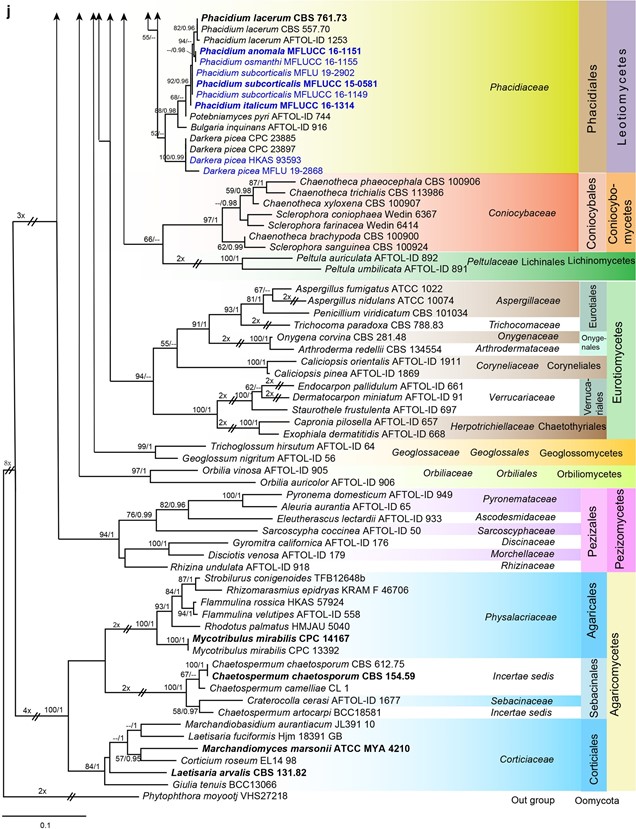
Fig. 7 (continued)
Species
