Cytospora salicicola C. Norphanphoun, Bulgakov & K.D. Hyde, sp. nov, Index Fungorum number: IF 551803
Etymology: Named after the host genus on which the fungus occurs.
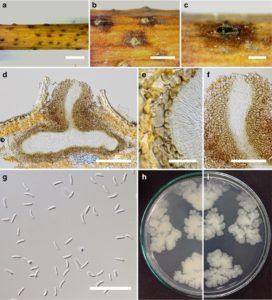
Pathogen causing dieback of twigs and branches of Salix alba L. Conidiomata 500–300μm diam. (x =400×350μm,
n =10), pycnidial, solitary, immersed in host tissue, unilocular, dark brown, ostiolate. Ostiole 150–40 μm diam. (x = 145 × 40μm, n = 10), at the same level as the disc surface. Peridium comprising a few to several layers of cell of textura angularis, with inner most layer thin, hyaline, outer layer brown to dark brown. Conidiophores reduced to conidiogenous cells. Conidiogenous cells blastic, enteroblastic phialidic, formed from the inner most layer of pycnidial wall, hyaline, smooth. Conidia (3.4–) 4.3–5.3 ×0.7– 0.8 (–1) μm (x =4.3×0.8μm, n = 30), unicellular, allantoid to subcylindrical, hyaline, smooth-walled. Sexual morph Undetermined.
Culture characteristics: Colonies on PDA, reaching 3.5 cm diam. after 10 days at 25 °C, producing dense mycelium, circular, rough margin white, after 5 days, flat or effuse on the surface, without aerial mycelium.
Material examined: RUSSIA, Rostov Region, Krasnosulinsky District, Donskoye forestry, riparian forest, on dead twigs and branches of Salix alba L. (Salicaceae), 21 May 2014, T.S. Bulgakov (MFLU 14–0785, holotype; PDD, isotype); ex-type-living cultures, MFLUCC 14–1052, ICMP.
Notes: Cytospora salicicola belongs in Valsaceae based on morphology and phylogeny. The new species has immersed, uniloculate conidiomata, with a single ostiole and shares common walls with the host tissue. Cytospora salicicola is most similar to C. schulzeri Sacc. & P. Syd. in conidia size
Fig. 1 Cytospora salicicola (holotype) a Appearance of fruiting bodies in wood b Fruiting bodies on substrate c Surface of fruiting bodies d Cross section of the conidioma e Peridium f Ostiole of conidioma g Conidia h–g Colonies on PDA (P from below). Scale bars: a = 2 mm, b–c = 1 mm, d = 100 μm, e = 10μm, f = 50 μm, and g = 20 μm.