Coronophorales Nannf., Nova Acta R. Soc. Scient. upsal., Ser. 4 8(no. 2): 54 (1932)
MycoBank number: MB 501516; Index Fungorum number: IF 501516; Facesoffungi number: FoF 06517;
= Melanosporales N. Zhang & M. Blackw., Mycol. Res. 111(5): 531 (2007)
Members of Coronophorales have a worldwide distribution as saprobes on wood (Mugambi & Huhndorf 2010, Maharachchikumbura et al. 2016b, Nannfeldt 1932). Most species in this order have superficial ascomata, sometimes with a subiculum (Mugambi & Huhndorf 2010). The quellkörper is a conical structure sometimes present in the centrum of the cupulate or collapsed ascomata and Munk pores are unique structures. These are rings of thickening between cells and are important characters in peridium cells (Huang et al. 2019, Mugambi & Huhndorf 2010). Coronophorales is a poorly studied group and was considered as synonym of Sordariales or Nitschkiaceae (Barr 1990b, Müller & von Arx 1973, Nannfeldt 1932, 1975, Nannfeldt & Santesson 1975, Subramanian & Sekar 1990). Huhndorf et al. (2004a) used LSU sequence data to show that Coronophorales is an independent member and lineage within Hypocreales, Hypocreomycetidae. Multi-gene analyses were used to demonstrate the position of Coronophorales which accommodates Bertiaceae, Ceratostomataceae, Chaetosphaerellaceae, Coronophoraceae, Nitschkiaceae and Scortechiniaceae (Hongsanan et al. 2017, Huang et al. 2019, Hyde et al. 2017a, Maharachchikumbura et al. 2015, Mugambi & Huhndorf 2010). However, Parasympodiellales clusters as a sister group for Scortechiniaceae in our phylogenetic analysis (Fig. 11). The divergence time for Coronophorales has been estimated as 192 MYA (Fig. 2). Currently there are six families and 46 genera in this order (this paper).
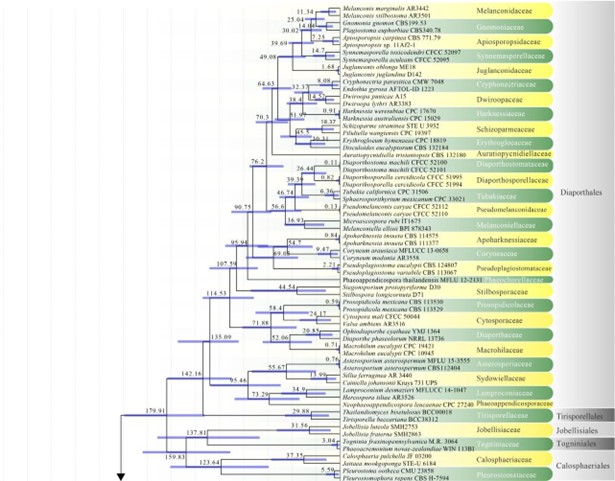
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
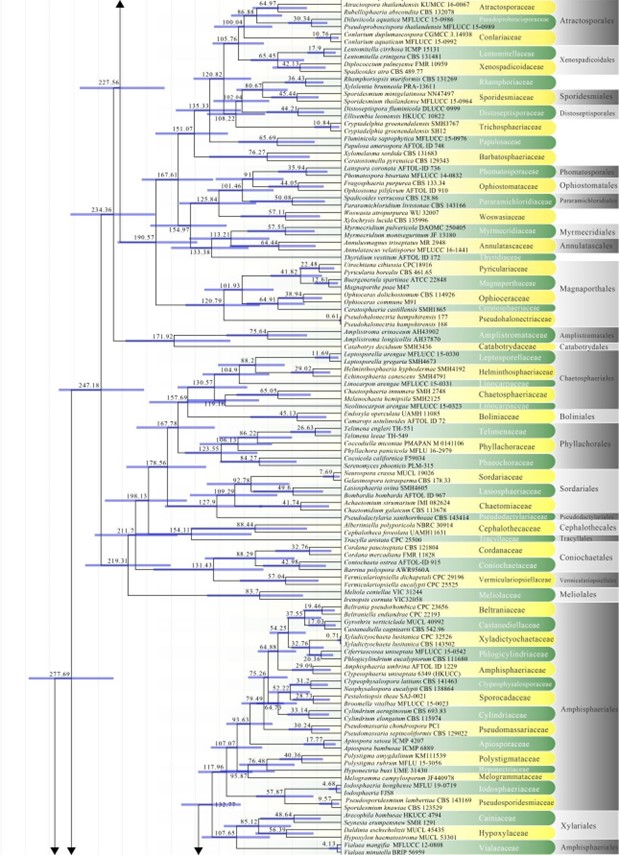
Figure 2 – Continued.
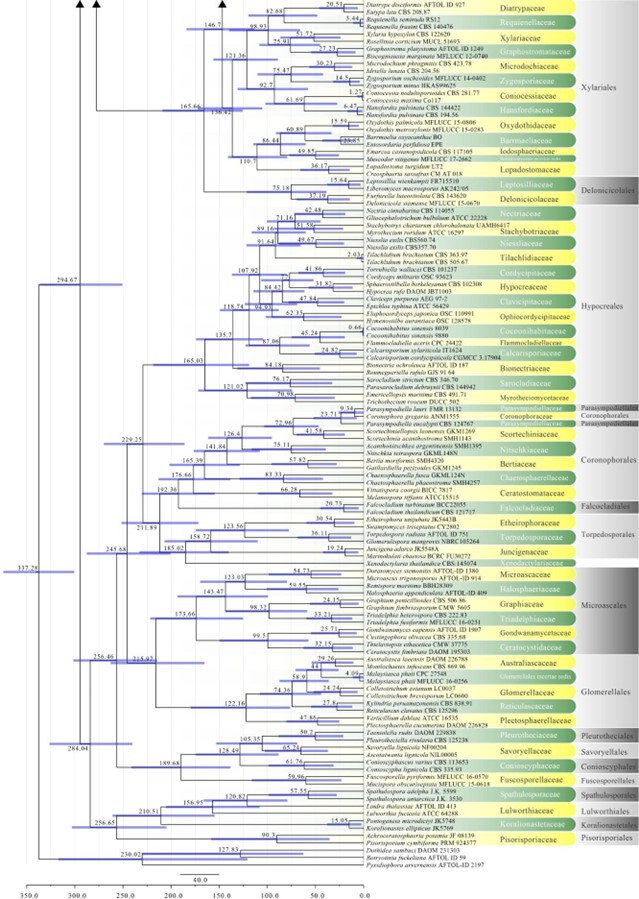
Figure 2 – Continued.
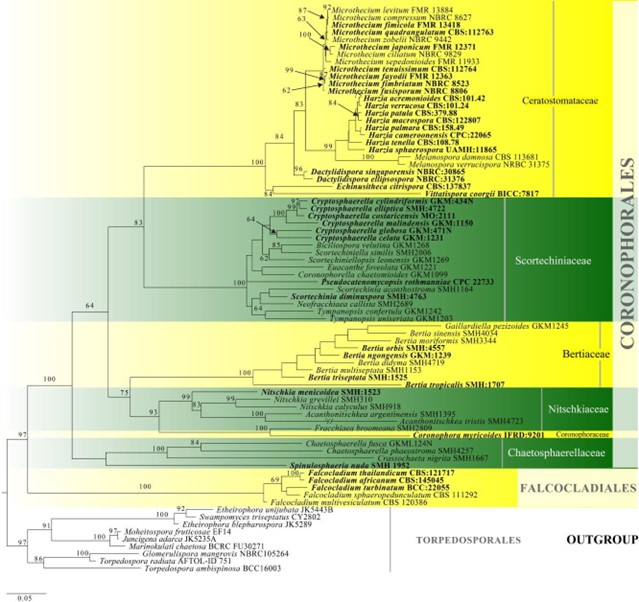
Figure 11 – Phylogram generated from maximum likelihood analysis based on combined LSU, tef1 and ITS sequence data for Coronophorales. Related sequences are referred to Hongsanan et al. (2017). Seventy-nine strains are included in the combined analyses which comprised 2211 characters (1044 characters for LSU, 627 characters for tef1, 540 characters for ITS) after alignment. Members of Torpedosporales are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. The best RaxML tree with a final likelihood value of -23354.525618 is presented. Estimated base frequencies were as follows: A = 0.228019, C = 0.288494, G = 0.281955, T = 0.201533; substitution rates AC = 1.194725, AG = 2.787601, AT = 1.703067, CG = 0.955075, CT = 3.781057, GT = 1.000000; gamma distribution shape parameter a = 0.736138. Bootstrap support values for ML greater than 50% are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
Families
Bertiaceae
Coronophoraceae
Scortechiniaceae
