Conlariaceae H. Zhang, K.D. Hyde & Maharachch., Fungal Divers. 85: 90 (2017)
MycoBank number: MB 553758; Index Fungorum number: IF 553758; Facesoffungi number: FoF 03336; 7 species.
Saprobic on submerged decaying wood in freshwater or grow on soil in terrestrial habitats. Sexual morph: Ascomata perithecioid, gregarious, coriaceous or membranous, immersed to erumpent or superficial, dark brown to black, globose to subglobose, smooth, ostiolate. Neck short or elongated, cylindrical, straight or slightly flexuous. Peridium composed of several layers of cells of textura angularis. Paraphyses cylindrical or globose, hyaline, septate. Asci 8-spored, unitunicate, cylindrical, short pedicellate, apically rounded, with a distinct, refractive, massive, barrel-shaped, apical ring or lacking apical structures. Ascospores uni- to bi-seriate, hyaline, fusiform or ellipsoidal-fusiform, straight or slightly curved, aseptate to multi-septate, guttulate, thin- or thick-walled, with or without appendages or appendages at one or both ends or surrounded by an irregular gelatinous sheath. Asexual morph: Hyphomycetous. Colonies dark brown to black. Mycelium mostly immersed, consisting of branched, septate, thin-walled, smooth, pale brown to brown hyphae. Conidiophores micronematous or semi-macronematous, mononematous, septate or aseptate, unbranched or irregularly branched, straight or flexuous, hyaline, becoming brown when old. Conidiogenous cells holoblastic, determinate, doliiform, cylindrical. Conidia brown, muriform, irregularly globose or subglobose, septate, constricted at the septa.
Type genus – Conlarium F. Liu & L. Cai
Notes – Conlariaceae was introduced in a new order Atractosporales by Zhang et al. (2017a) for a single genus Conlarium which comprised C. aquaticum and C. dupliciascosporum. Asexual morphs of the family were found in the culture of C. dupliciascosporum (Liu et al. 2012), on natural decaying submerged wood (Zhang et al. 2017a) and dead wood or soil in terrestrial habitat (Phookamsak et al. 2019, Xie et al. 2019). Another genus Riomyces is included in the family based on our phylogenetic analyses (Fig. 3), and previous study (Luo et al. 2019). The phylogeny and morphology warranted a new family (Zhang et al. 2017a).
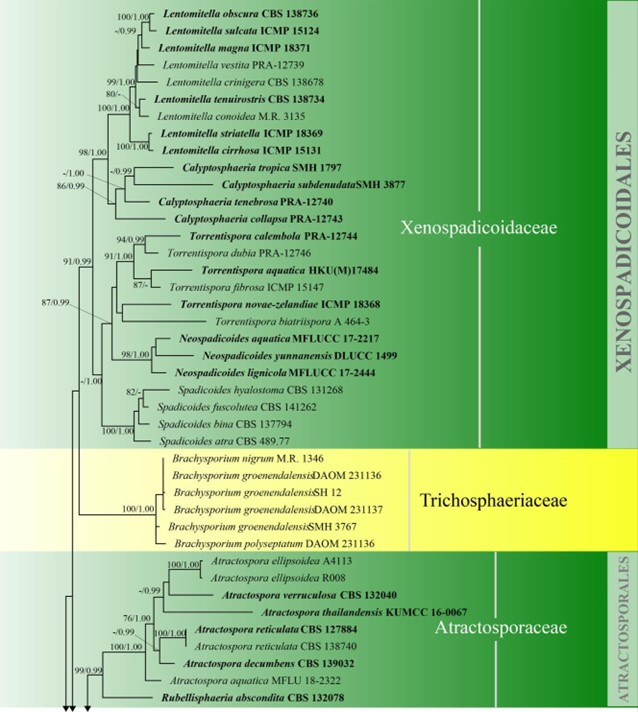
Figure 3 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and rpb2 sequence data of Diaporthomycetidae. One hundred and ninety-three strains are included in the combined analyses which comprised 3545 characters (859 characters for LSU, 972 characters for SSU, 659 characters for ITS) after alignment. Single gene analyses were carried out and the topology of each tree had clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 68207.368884 is presented. Estimated base frequencies were as follows: A = 0.248206, C = 0.241993, G = 0.285500, T = 0.224301; substitution rates AC = 1.369088, AG = 2.887040, AT = 1.413053, CG = 1.152137, CT = 6.303994, GT = 1.000000; gamma distribution shape parameter a = 0.315782. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. The tree is rooted with Diatrype disciformis (AFTOL-ID 927). Ex-type strains are in bold. The newly generated sequences are indicated in blue.
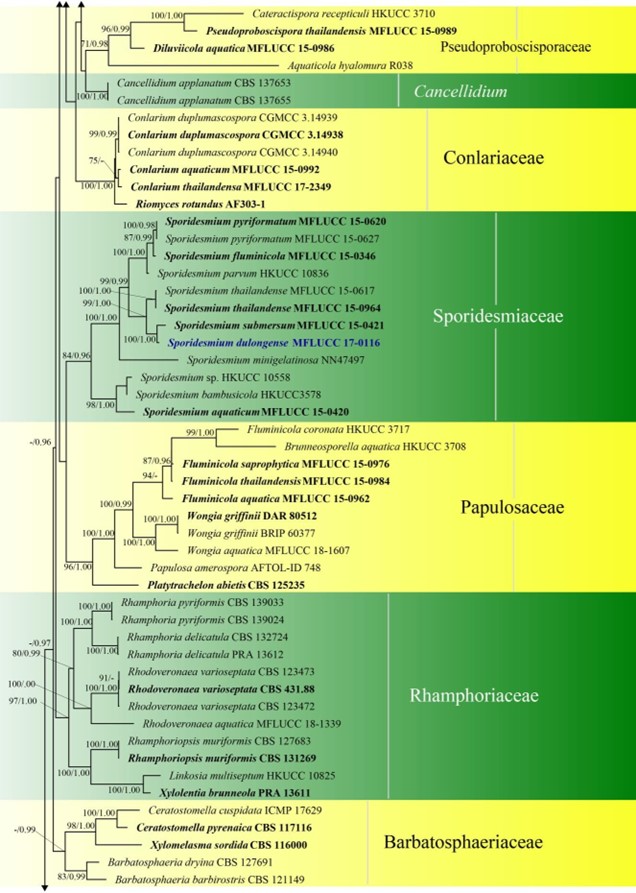
Figure 3 – Continued.
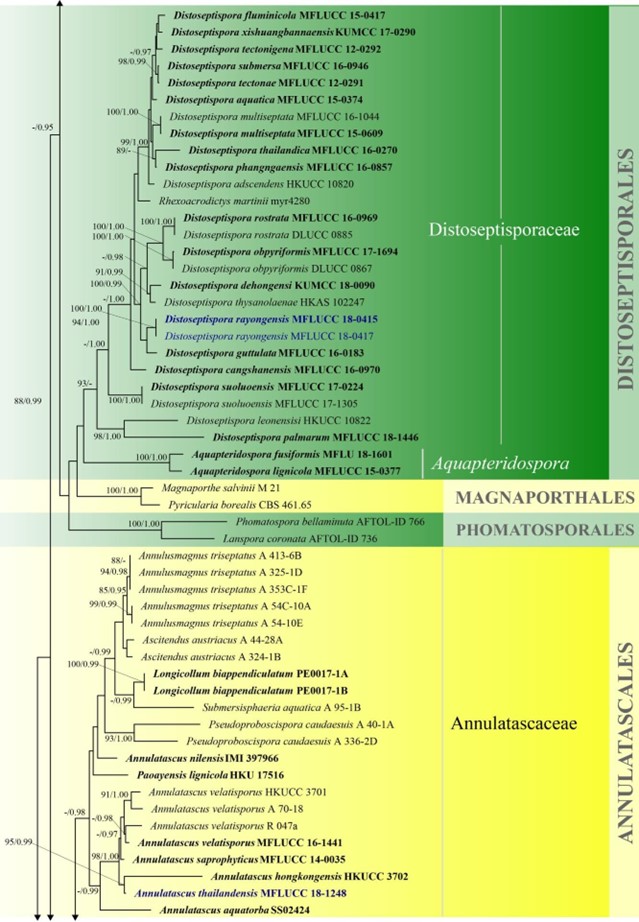
Figure 3 – Continued.
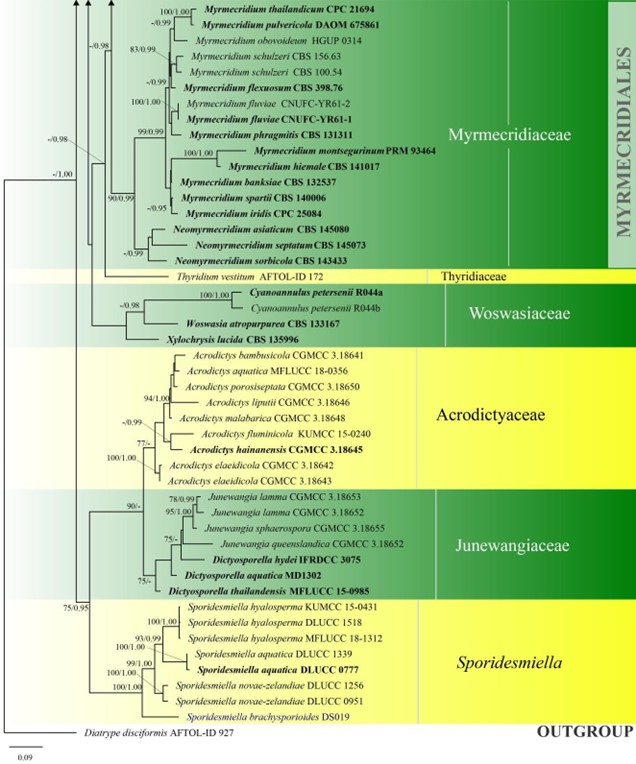
Figure 3 – Continued.
