Colletotrichum dracaenophilum D.F. Farr & M.E. Palm, Mycol. Res. 110: 1401. 2006. (SBRBR1) (Ruvi)
MycoBank number: MB 510231; Index Fungorum number: IF 510231; Facesoffungi number: FoF 10776; Fig. **
Saprobic on dead stems of Dracaena sp. Sexual morph: Undetermined. Asexual morph: Conidiomata, solitary or aggregate. Conidiophores hyaline to medium brown, smooth-walled, verruculose towards the tip, 50–130 μm long, 2–5-septate, base cylindrical, sometimes slightly inflated, 5–8 μm diam, tip ± acute, often ending in a conidiogenous opening. Conidiomata, conidiophores and setae formed directly on hyphae. Conidiogenous cells hyaline to medium brown, smooth-walled, cylindrical, the upper part sometimes surrounded by a gelatinous sheath, 12–24 × 3–6 μm, opening 1–2 μm diam, collarette 0.5 μm long, periclinal thickening distinct. Conidia hyaline, smooth-walled, aseptate, the apex rounded, the base rounded to truncate, 15–20 × 4–5 μm, sometimes tapering to the base and sometimes slightly curved. Appressoria not formed.
Material examined – Thailand, Ratchaburi Province, Pak Tho District, on Dracaena sp., 2 September 2017, Saranyaphat Boonmee, SBRBR1 (MFLU ***, holotype), ex-type living culture, MFLUCC 18–0489.
Host – Dracaena sanderiana (Damm et al. 2018), Dracaena sp.—(This study).
Distribution – Bulgaria, China, USA (Damm et al. 2018), Thailand—(This study).
GenBank accession numbers – MFLU **; ITS: ****; TUB2: waiting; ACT: waiting CHS waiting GAPDH: waiting
Notes – The first report of Colletotrichum dracaenophilum was described by Farr et al. 2006 as a stem pathogen of Dracaena sanderiana in California, USA. Colletotrichum dracaenophilum species have been report only from D. sanderiana in Australia, Bulgaria, China, Egypt and Florida (USA), and on D. braunii in Brazil (Farr et al. 2006, Bobev et al. 2008, Sharma et al. 2014, Macedo & Barreto 2016, Morsy & Elshahawy 2016, Shivas et al. 2016). This species belongs to the C. dracaenophilum species complex. The closest matches with the sequences of all loci of the ex-type strain CBS 118199 in GenBank are all those of the C. dracaenophilum strains included in this study. In a BLASTn search of GenBank, the ITS sequence had ***% similarity (**% nucleotide differences), while the act sequence had **% similarity (** nucleotide differences in 297 nucleotides). Thus, our strain is introduced as a new geological record of C. dracaenophilum from Dracaena sp. in Thailand.
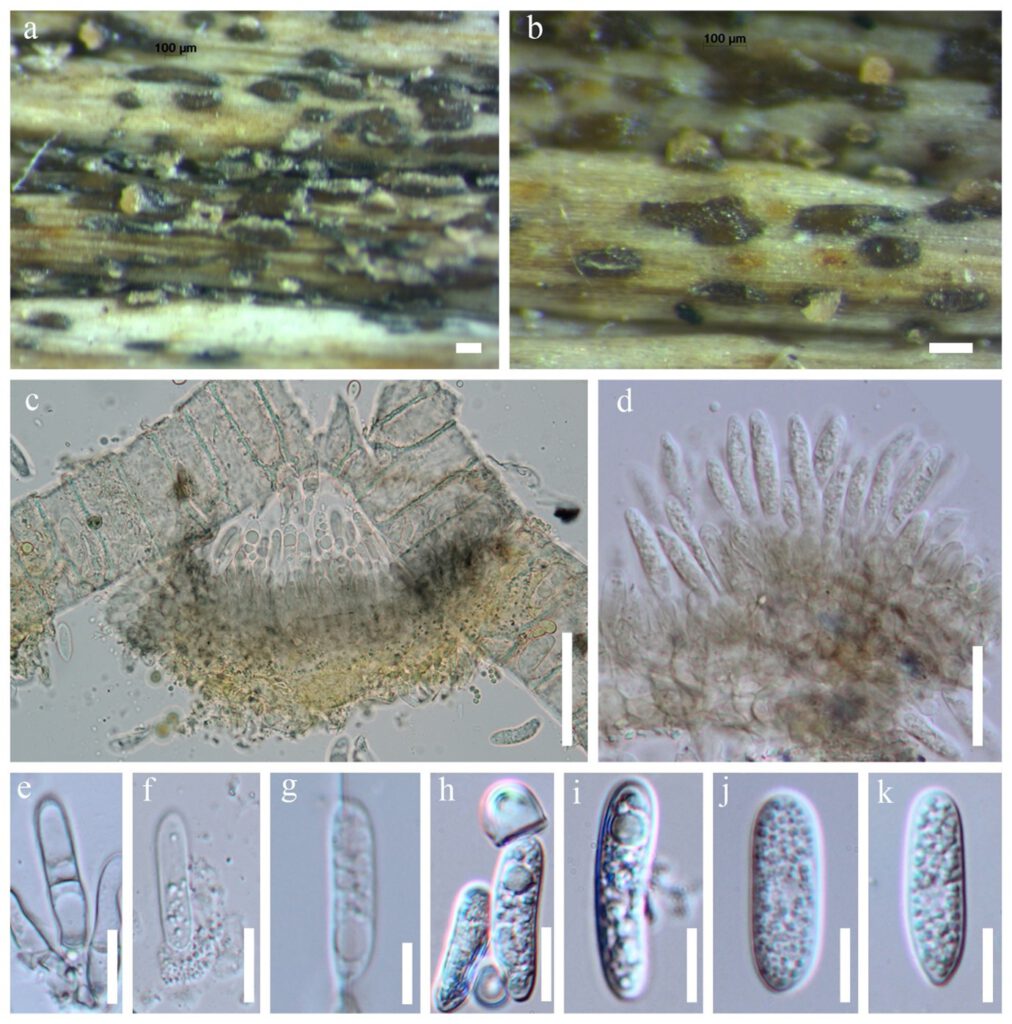
Figure** – Colletotrichum dracaenophilum (MFLU **** SBRBR1). a, b Fruiting body on dead leaf of Dracaena sp. c–e Conidiogenous cells f–k Conidia. Scale bars: a, b 100 μm, c = 40 μm, d = 20 μm, e–k = 10 μm.
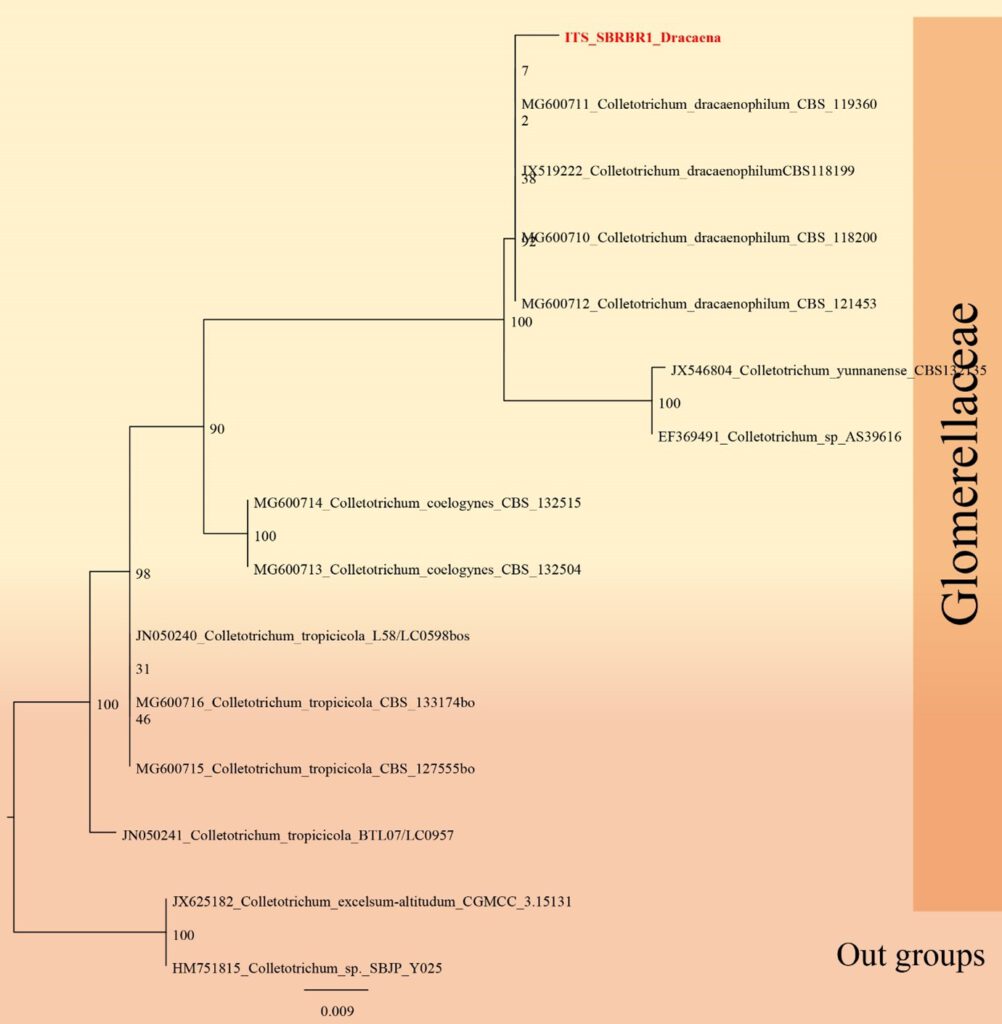
Figure**– Phylogram generated from RAxML analysis based on ITS sequence data of selected Glomerellaceae isolates. Related sequences were obtained from GenBank. Twenty-four taxa are included in the analyses, which comprise *** characters including gaps. The tree is rooted to Colletotrichum excelsum-altitudum. The best scoring RAxML tree with a final likelihood value of − 7281.602128 is presented. The matrix had 613 distinct alignment patterns, with 25.61% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.244440, C = 0.238073, G = 0.310001, T = 0.207486; substitution rates AC = 1.017532, AG = 2.172618, AT = 1.516851, CG = 1.265405, CT = 8.040081, GT = 1.000000; gamma distribution shape parameter α = ***. Maximum likelihood bootstrap support values ≥ 75% (BT) are given at the nodes as ML. The scale bar indicates 0.07 changes. The isolates obtained in this study are in red and extypes taxa are in black bold
