Chloridium Link, Mag. Gesell. naturf. Freunde, Berlin 3(1–2): 13 (1809)
MycoBank number: MB 7624; Index Fungorum number: IF 7624; Facesoffungi number: FoF 13052; 28 morphological species (Species Fungorum 2020), 5 species with sequence data.
Type species – Chloridium viride Link
Notes – Chloridium is characterised by mononematous or macronematous conidiophores, monophialidic or polyphialidic conidiogenous cells, with a funnel-shaped collarette and hyaline to brown, eguttulate or guttulate, appendaged or non-appendaged conidia (Link 1809, Seifert et al. 2011, Wei et al. 2018). Réblová et al. (2016b) pointed out that the generic type of Chloridium, C. viride is congeneric with Melanopsammella inaequalis, the type of Melanopsammella (Réblová & Winka 2000, Fernández et al. 2006, Crous et al. 2012e). Hughes (1951c) revealed that the generic type of Gonytrichum, G. caesium, is the asexual morph of Melanopsammella inaequalis. By considering the above facts, Réblová et al. (2016b) proposed that Gonytrichum and Melanopsammella are synonyms of Chloridium. Hence, the widely used generic name Chloridium was conserved over Gonytrichum and Melanopsammella (Réblová et al. 2016b, Wei et al. 2018). Wijayawardene et al. (2018a) incorrectly treated Chloridium, Gonytrichum and Melanopsammella as three distinct genera. Here we follow Réblová et al. (2016b) which is also confirmed in our phylogenetic analysis of combined ITS and LSU sequence data (Fig. 8). Furthermore, Chloridium species seems to be polyphyletic within Chaetosphaeriaceae (Fig. 8). We introduce a new species of Chloridium in this entry.

Figure 8 – Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data of Chaetosphaeriales and Tracyllalales taxa. Ninety-six strains are included in the combined analyses which comprised 1695 characters (1081 characters for LSU, 614 characters for ITS) after alignment. Neurospora crassa MUCL 19026 and Gelasinospora tetrasperma CBS 178.33 (Sordariaceae, Sordariales) are used as outgroup taxa. Single gene analyses were carried out and the phylogenies were similar in topology and clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 23777.689886 is presented. Estimated base frequencies were as follows: A = 0.231060, C = 0.264793, G = 0.308265, T = 0.195882; substitution rates AC = 1.388486, AG = 1.836207, AT = 1.649563, CG = 0.971659, CT = 6.316962, GT = 1.000000; gamma distribution shape parameter a = 0.460297. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. Ex-type strains are in bold. The newly generated sequences are indicated in blue.
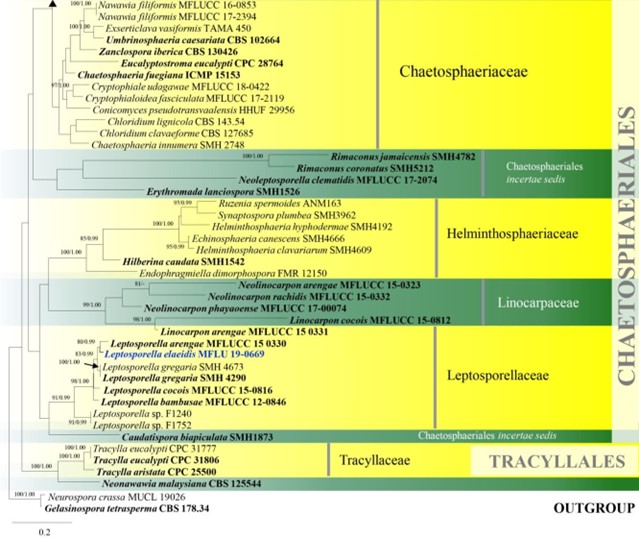
Figure 8 – Continued
Species
Chloridium viride
