Camporesiomyces mali D.P. Wei & K.D. Hyde, sp. nov.
MycoBank number: MB 557016; Index Fungorum number: IF 557016; Facesoffungi number: FoF 07075; Fig. 88
Etymology: The specific epithet “mali” refers to the host genus Malus.
Holotype: KUN-HKAS 102565
Saprobic on decaying branches of Malus halliana Koehne. Sexual morph Ascomata 200–300 μm (x̄ = 231 μm, n = 5), subimmersed, scattered, black, nearly globose, multi-loculate, ostiolate, setaceous. Setae 15–20 × 1–2 μm (x̄ = 17 × 1.5 μm, n = 5), short, dark brown to black, aseptate, attenuated toward the apex. Peridium 15–50 μm (x̄ = 23 μm, n = 20), comprising dark brown to pale-brown, irregular in thickness, thin-walled cells of textura angularis. Hamathecium comprising inconspicuous, filiform, hyaline pseudoparaphyses. Asci 50–100 × 10–25 μm ( x̄ = 70 × 14 μm, n = 10), 8-spored, bitunicate, broadly clavate, thickened at apex, without ocular chamber. Ascospores 50–75 × 3–5 μm ( x̄ = 62 × 4 μm, n = 30), hyaline, cylindrical, slightly curved, 10–12-septate, ends remaining cone-shaped, smooth-walled, guttulate when young.
Culture characteristics: Culture was obtained from germinated ascospores. Colonies slowly growing on PDA, brown, slightly raised, dense, with irregular margin, reaching 2 mm in 9 days at 25 °C.
Material examined: CHINA, Yunnan Province, Kunming City, Kunming Institute of Botany, on decaying wood branches of Malus halliana (Rosaceae), 19 August 2018, D.P. Wei, KIB1903 (KUN-HKAS 102565, holotype), ex-type living culture, KUMCC 19-0216.
GenBank numbers: ITS =MN792813, LSU = MN792811, TEF1-α = MN794018.
Notes: Our isolate has close affinity with Acanthostigma patagonicum (BBB MVB 573) and Helicoma vaccinii (CBS 216.90) by forming a distinct, strongly supported clade as basal to the remaining taxa in Helicosporium (100% ML, Fig. 89). Our isolate resembles A. patagonicum in having dark brown setae, clavate asci and inconspicuous pseudoparaphyses. But our isolate can be distinguished from A. patagonicum in having narrowly fusiform, symmetric ascospores, in contrast to elongate fusiform ascospores, with asymmetrical ends in A. patagonicum (Sanchez et al. 2012). Helicosporium vaccinii is only known from its asexual morph (Carris 1989). A comparison of ITS sequences between our isolate and A. patagonicum show 24 bp difference and 3 gaps within 330 bp nucleotides. When ITS sequences are compared with H. vaccinii, there are 52 bp difference and 17 gaps within 504 bp nucleotides. Thereby, we determined our isolate to be a novel species in the new genus Camporesiomyces.
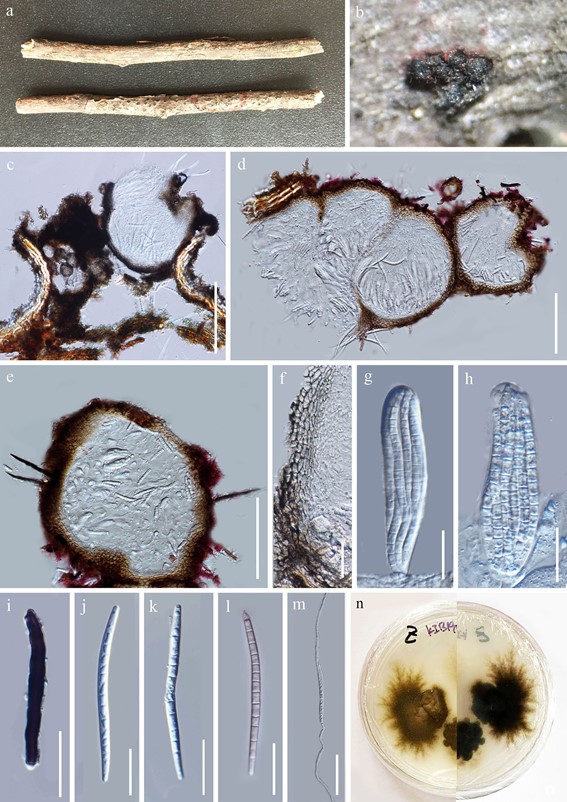
Fig. 88 Camporesiomyces mali (KUN-HKAS 102565, holo- type). a Substrate. b Ascomata on substrate. c, d Transverse section of ascomata. e Setae on ascoma. f Peridium. g, h Asci. i Seta. j–l Ascospores. m Germinated ascospore. n Colony on PDA from above and below. Scale bars: c = 150 µm, d, e = 100 µm, m = 50 µm, f–l = 20 µm
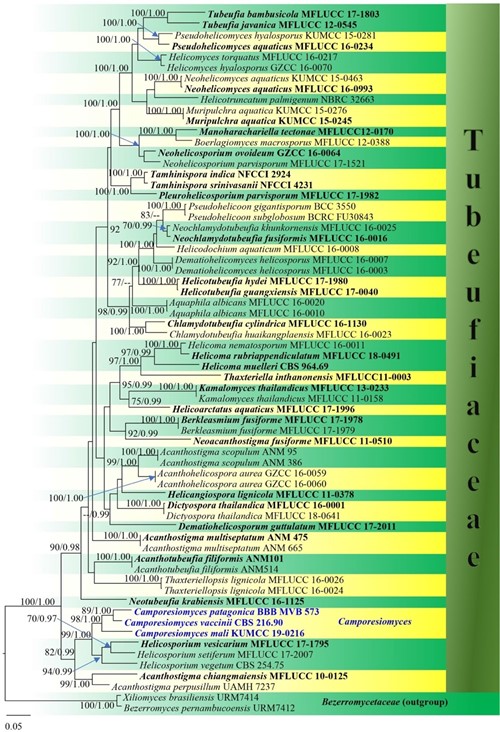
Fig. 89 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, TEF1-α, and RPB2 sequence dataset representing taxa in family Tubeufiaceae (Tubeufiales). Related sequences are taken from Lu et al. (2018). Sixty-six strains are included in the combined analyses which comprise 3219 characters (828 characters for LSU, 509 characters for ITS, 837 characters for TEF1-α and 1045 characters for RPB2) after alignment. The best scoring RAxML tree with a final likelihood value of − 31516.131561 is presented. The matrix had 1311 distinct alignment patterns, with 25.67% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243290, C = 0.257008, G = 0.265413, T = 0.234289; substitution rates: AC = 0.995531, AG = 4.500491, AT = 2.148310, CG = 0.787501, CT = 8.890762, GT = 1.000000; gamma distribution shape parameter α = 0.791557. Bootstrap support values for ML equal to or greater than 70% and BYPP equal to or greater than 0.95 are given above the nodes. Newly generated sequences are in blue bold and type species are in black bold. Xiliomyces brasiliensis (URM7414) and Bezerromyces pernambucoensis (URM7412) in family Bezerromycetaceae (Tubeufiales) were used as the outgroup taxa
