Calosphaeriales M.E. Barr, Mycologia 75(1): 11 (1983)
MycoBank number: MB 90463; Index Fungorum number: IF 90463; Facesoffungi number: FoF 01133;
Members of Calosphaeriaceae and Pleurostomataceae are accommodated in Calosphaeriales and are characterized by brown to black ascomata with reniform to clavate asci, phialidic conidiophores and ellipsoid to allantoid conidia (Maharachchikumbura et al. 2016b, Huang et al. 2019). Some species are plant pathogens (Barr 1983, Damm et al. 2008, Trouillas et al. 2012). In previous phylogenetic analyses, species of Calosphaeriales were closely related to Jobellisiales and Togniniales (Maharachchikumbura et al. 2015, 2016b, Hongsanan et al. 2017). Calosphaeriales comprises seven genera within the two families Calosphaeriaceae and Pleurostomataceae (Huang et al. 2019). The monotypic genera, Kacosphaeria and Sulcatistroma are accepted in Calosphaeriales based on the small, clavate asci with allantoid to ellipsoid, hyaline ascospores similar to Calosphaeria (Huang et al. 2019). The divergence time for Calosphaeriales has been estimated as 160 MYA (Fig. 2). Currently there are two families and nine genera in this order (this paper).
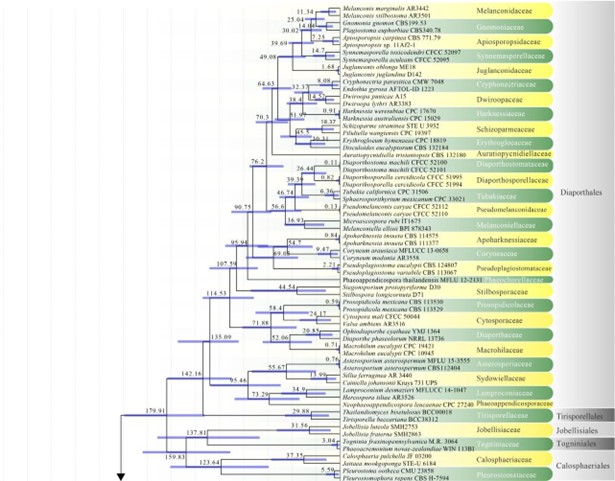
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
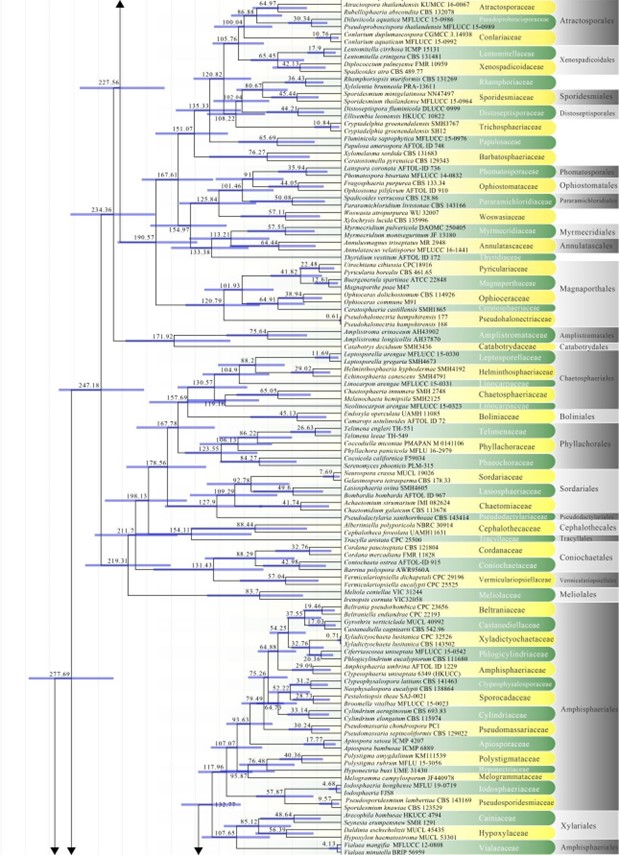
Figure 2 – Continued.
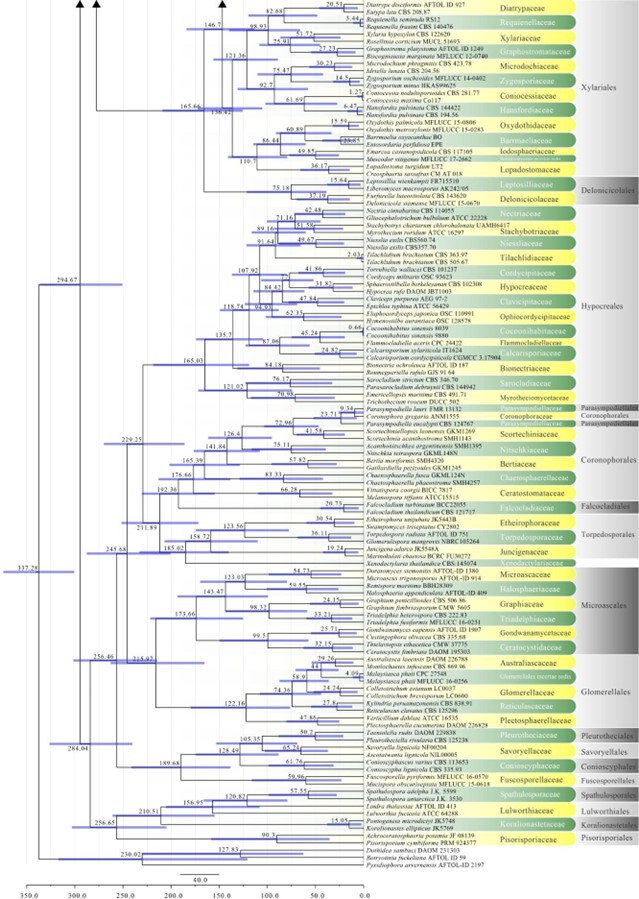
Figure 2 – Continued.
Families
