Bartalinia caryotae Senan., Kular. & KD Hyde, in Jayawardena et al., Fungal Diversity 117: 148 (2023) [2022]
Index Fungorum number: IF 558407; MycoBank number: MB 558407; Facesoffungi number: FoF 10699; Fig. 1
Etymology – Species epithet based on the host genus.
Holotype – HKAS115853
Associated with leaf–tip spots of Caryota sp. Sexual morph: Not observed. Asexual morph: Coelomycetous. Conidiomata 55–75 × 115–140 μm (x̅ = 62 × 120 μm, n = 15), pycnidial, superficial or rarely erumpent, solitary, scattered, dark brown, uniloculate, conical, glabrous, ostiolate, without a papilla. Conidiomata wall 3–8 μm wide (x̅ = 6 μm, n = 5), comprising several layers of brown, pseudoparenchymatous cells of textura angularis, paler towards the inner layers. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–7 × 1.5–4 μm (x̅ = 5 × 3 μm, n = 10), enteroblastic, annellidic, integrated, hyaline to subhyaline, ampulliform to subcylindrical, or obclavate, aseptate, thin- and smooth-walled. Conidia 17.5–19 × 5–7 μm (x̅ = 18 × 6 μm, n = 20), subcylindrical, hyaline when immature, olivaceous when mature, straight to slightly curved, 4-septate, constricted at septa, smooth, with longest cell second from base, bearing appendages at both ends; basal cell 2–4 μm long, conical, hyaline, to pale brown, paler than middle cells; second cell from base 5–8 μm long, pale brown; third cell 3–5 μm long, pale brown; fourth cell 3–5 μm long, pale brown; apical cell 2–3 μm long, conical, hyaline, smooth-walled with a tubular, flexuous, divergently 3–4, unbranched, 20–30 μm long, tubular appendages arising from the tip of apical cell, with a basal 10–25 μm long, centric to eccentrically located, unbranched, flexuous, tubular to filiform appendage.
Culture characteristics – Colonies on PDA reaching 2 cm diam. after 5 days at 16 °C, flattened, circular, smooth margin, white with off-white aerial mycelia; reverse cream.
Material examined – China, Guangdong Province, Shenzhen, Nanshan District, Mountain Yangtai Forest Park, 22°39′21.26″N 113°57′18.53″E, living leaves of Caryota sp (Arecaceae), 4 October 2019, ND Kularathnage, NDK 24, (HKAS115853, holotype), ex-type culture, ZHKUCC 21-0093; ibid., 15 September 2018, IC. Senanayake, SI 43, (ZHKU 21-0005, paratype), ex-type culture, ZHKUCC 21-0094.
GenBank accession numbers – ZHKUCC 21-0093: ITS = MZ520792, LSU = MZ520794; ZHKUCC 21-0094: ITS = MZ520793, LSU = MZ520795.
Notes – In the combined ITS and LSU gene analysis, Bartalinia caryotae formed a distinct clade with high bootstrap support (MP/BI=100/1), forming a basal to B. kevinhydei (Fig. 1). Bartalinia caryotae is morphologically different from Bartalinia kevinhydei in having small, conical conidiomata with large, 3-4 unbranched apical appendages (Liu et al. 2019). Therefore, we introduce Bartalinia caryotae as a novel taxon from the leaves of Caryota species and Bartalinia species are rarely reported on monocotyledon plants. Our species forms superficial to erumpent conidiomata on leaves.
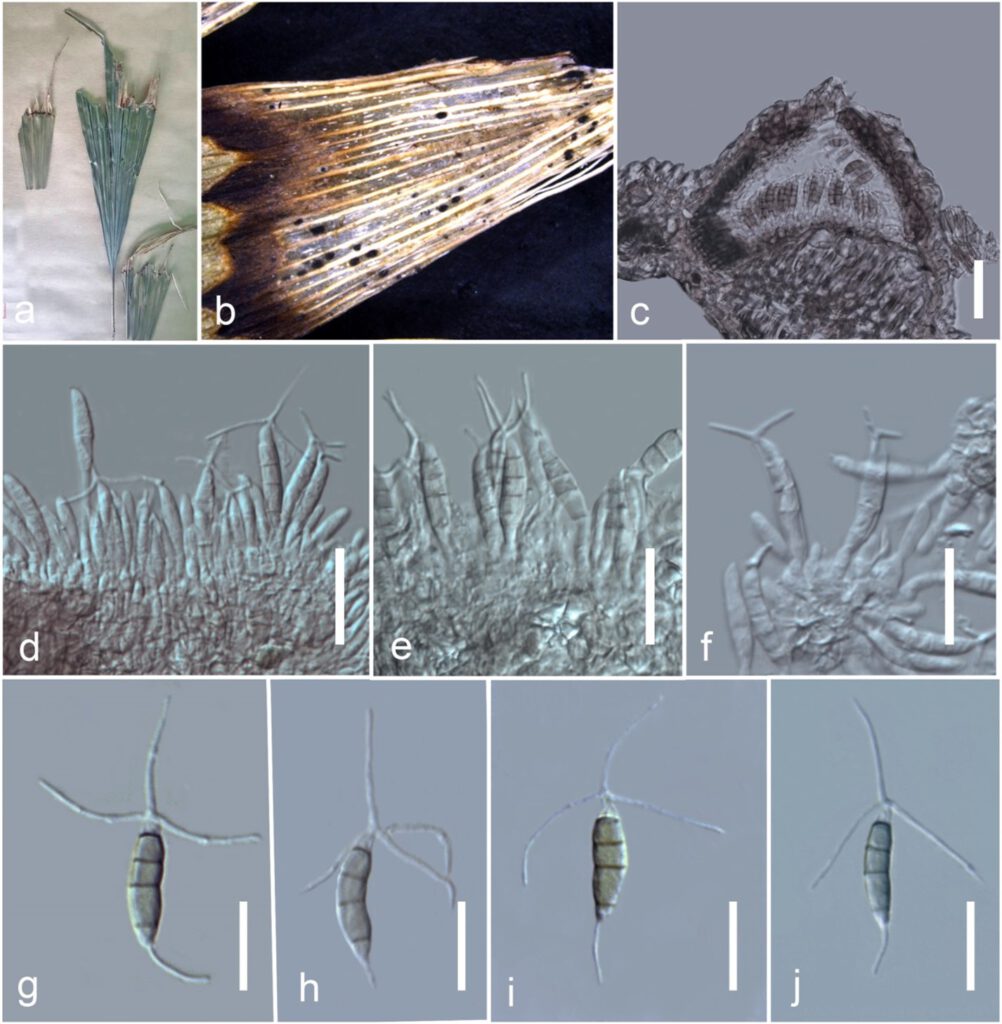
Fig 1. Bartalinia caryotae (HKAS115853, holotype). a Specimen. b Conidiomata on substrate. c Cross section of conidiomata. d–f Conidia attached to conidiogeneous cells. g–j Conidia. Scale bars: c = 30 µm, d–f = 20 µm, g–j = 15 µm.
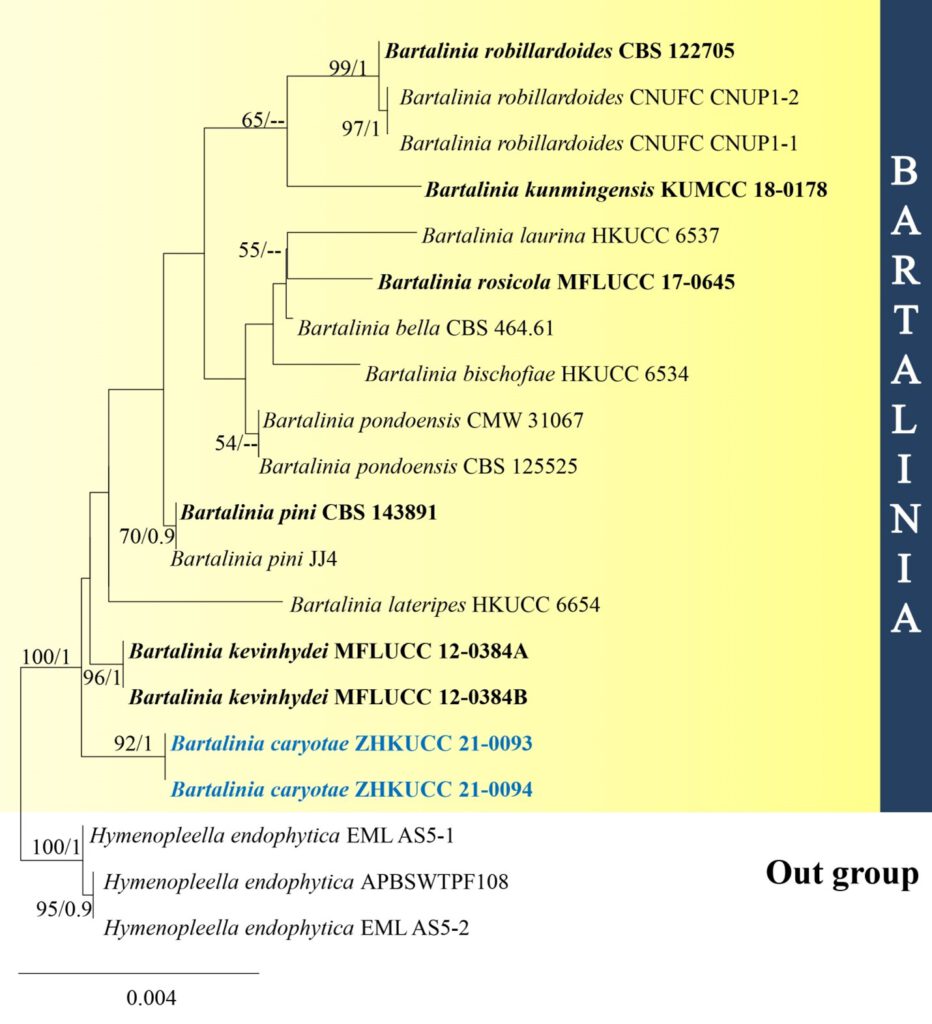
Fig xx. Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data. Twenty taxa were included in the combined analyses, which comprised 1479 characters (LSU = 897 bp, ITS = 582 bp) after alignment. The best scoring RAxML tree with a final likelihood value of -2522.918035 is presented. The matrix had 1494 distinct patterns, with 19.52% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.268501, C = 0.209785, G = 0.258291, T = 0.263423; substitution rates: AC = 1.249704, AG = 3.871912, AT = 1.829117, CG = 2.581099, CT = 6.657265, GT = 1.000000; gamma distribution shape parameter α = 0.020137. Bootstrap support values for maximum likelihood (ML) equal to or greater than 50% and clade credibility values greater than 0.90 from Bayesian inference analysis are labelled at each node. Ex-type strains are in bold and the new isolate is indicated in blue bold. Hymenopleella endophytica (APBSWTPF108, EML AS5-1, EML AS5-2) were used as the outgroup taxa.
