Azygosporus B. Huang & Y. Nie, gen. nov.
MycoBank number: MB 840849; Index Fungorum number: IF 840849; Facesoffungi number: FoF;
Etymology: Referring to produce azygospores.
Type species: Azygosporus parvus (Drechsler) B. Huang & Y. Nie.
Description: Mycelia colorless. Primary conidiophores simple, bearing single pri- mary conidia. Primary conidia forcibly discharged multinucleate, colourless, globose to subglobose, small, less than 22.5 µm. Producing only globose or subglobose replica- tive conidia, similar to and smaller than primary conidia. Azygospores formed in the middle region of the old hyphal segments. Mature azygospores colourless or yellowish, smooth, without thickening or less thickening (0.5–1.2 µm).
Notes: – Azygosporus is strongly supported as monophyletic and is distinguished from other Ancylistaceae lineages by the synapomorphy of azygospore production.
Therefore, we classify this lineage as a new genus, named Azygosporus gen. nov. Azy- gosporus currently contains only two members: C. parvus (= A. parvus) and A. macro- papillatus sp. nov. (Fig. 1). Morphologically, Azygosporus is most similar to Microcon- idiobolus, which forms small primary conidia (less than 22.5 µm) (Table 2). However, the synapomorphy of azygospore production clearly distinguishes Azygosporus from Microconidiobolus and other allied genera of the family.
Table 2. Morphological measurements of A. macropapillatus and other related species.
| Species | Growth rate (mm/d) at
21oC on PDA |
Diameter of mycelia
(µm) |
Primary conidio- phores (µm) | Primary conidia (µm) | Basal papilla (µm) | Resting spores (µm) | References |
| A. macro- papillatus | 5.7–7.7 | 3.0–7.5 | 37.0–150.0×5.0–
8.5 |
16.5–22.5×12.0–
19.0 |
7.5–10.0×5.0–
10.0 |
azygosporus, 25.0–
30.0×27.0– 34.0 |
This article |
| A. parvus | 1.5 | 1.4–8.0
(3.5–5) |
15.0–30.0×3.0–8.0 | 6.0–20.0×4.5–17.0 | 1.5–6.0×1.5–4.5 | azygosporus, 20.0–
25.0×8.0– 20.0 |
Drechsler 1962 |
| M.
nodosus |
7.1 | 3.5–6.5 | 30.0–50.0 | 17.0–22.0×13.0–
16.0 |
2.5–5.0×1.5–2.5 | chlamydo- sporus | Srinivasan and Thirumalachar 1967; King
1977 |
| M. paulus | 1.3–3.3 | 1.5–7.0
(4.0–5.0) |
15.0–30.0×3.5–7.0 | 5.0–19.0×4.0–14.0 | 2.0–7.0×1.0–5.0 | zygosporus,
10.0–15.0 |
Drechsler 1957 |
| M. ter- restris | 2.6 | 2.8–4.5 | 15.0–80.0
×3.0–5.0 |
8.0–12.0 | 2.0–4.0×1.5–2.0 | chlamydo- sporus | Srinivasan and Thirumalachar 1968a; King
1977 |
| N. lam- prauges | less than 5.0 | 3.0–8.0
(4.0–7.0) |
25.0–100.0 (25.0–
50.0)×4.0–8.0 (5.0–15.0) |
15.0–22.0×12.5–
20.0 |
2.5–7.0×1.5–4.0 | zygosporus, 12.0–18.0 | Drechsler 1953 |
| N. kuny-
ushanensis |
8.3–10.0 | 3.5–9.0 | 62.0–121.0×7.0–
12.0 |
15.0–21.0×13.0–
17.0 |
4.0–8.0×1.0–4.0 | zygosporus,
12.0–25.0 |
Nie et al. 2021 |
| N.
pachyzy- gosporus |
12.0 | 3.0–14.0 | 34.0–156.0×6.0–
12.0 |
15.5–23.0×11.0–
18.0 |
3.0–5.0×1.0–4.0 | zygosporus, 15.0–25.0 | Nie et al. 2018 |
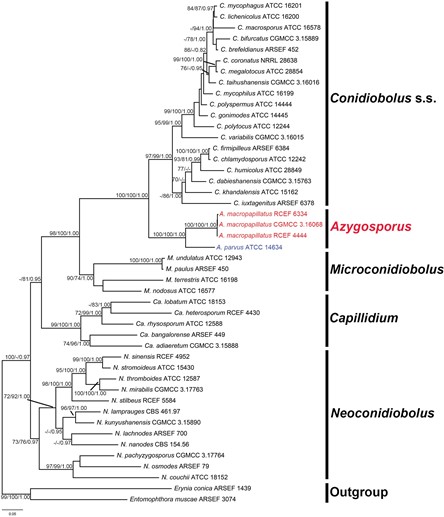
Figure 1. ML tree of Coniobolus s.l. using nrLSU + TEF1 + mtSSU sequences. Entomophthora muscae and Erynia conica are selected as outgroups. Support for each node is shown as MP bootstrap support/ ML bootstrap support/Bayesian posterior probability (MPBS/MLBS/BPP) for nodes with MPBS≧70%, MLBS≧70%) and BPP≧0.95. The new genus, Azyosporus, and new species, A. macropapillatus, are shown in red, and the new combination is shown in blue.
Species
