Atractosporales H. Zhang, K.D. Hyde & Maharachch., Fungal Divers. 85: 88 (2017)
MycoBank number: MB 553756; Index Fungorum number: IF 553756; Facesoffungi number: FoF 03333;
Atractosporales was introduced to resolve the taxonomic problems of annulatascaceae-like taxa previously placed in Annulatascaceae and Sordariomycetes genera incertae sedis. Three families (viz. Atractosporaceae, Conlariaceae, and Pseudoproboscisporaceae) were included in the order by Zhang et al. (2017a). The monophyly of this order is not well-supported in previous studies (Luo et al. 2019) and in this study. Luo et al. (2019) showed that Junewangiaceae and Cancellidium also clustered in Atractosporales and this agrees with our phylogenetic analyses, but with low bootstrap support (Fig. 3). We therefore retain Cancellidium (Tubaki 1975) as Sordariomycetes genera incertae sedis. This ambiguous group may be included in this order with larger taxon sampling in future phylogenetic analyses. The divergence time for Atractosporales has been estimated as 106 MYA (Fig. 2), which falls in the range for family status. The status of Atractosporales members may therefore need revision following further study. Currently there are three families and six genera in this order (this paper).
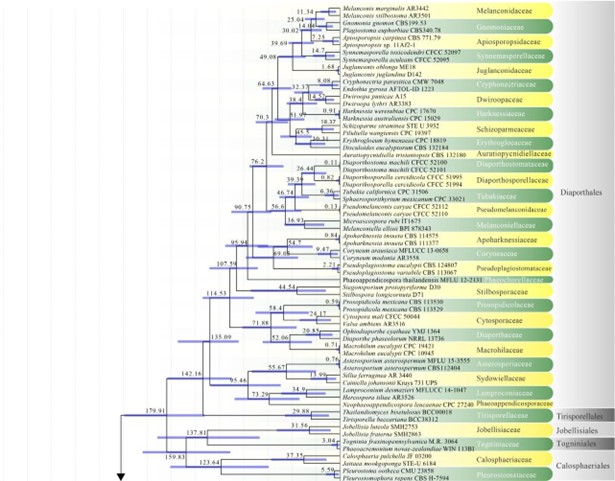
Figure 2 – The maximum clade credibility (MCC) tree, using the same dataset from Fig. 1. This analysis was performed in BEAST v1.10.2. The crown age of Sordariomycetes was set with Normal distribution, mean = 250, SD = 30, with 97.5% of CI = 308.8 MYA, and crown age of Dothideomycetes with Normal distribution mean = 360, SD = 20, with 97.5% of CI = 399 MYA. The substitution models were selected based on jModeltest2.1.1; GTR+I+G for LSU, rpb2 and SSU, and TrN+I+G for tef1 (the model TrN is not available in BEAUti 1.10.2, thus we used TN93). Lognormal distribution of rates was used during the analyses with uncorrelated relaxed clock model. The Yule process tree prior was used to model the speciation of nodes in the topology with a randomly generated starting tree. The analyses were performed for 100 million generations, with sampling parameters every 10000 generations. The effective sample sizes were checked in Tracer v.1.6 and the acceptable values are higher than 200. The first 20% representing the burn-in phase were discarded and the remaining trees were combined in LogCombiner 1.10.2., summarized data and estimated in TreeAnnotator 1.10.2. Bars correspond to the 95% highest posterior density (HPD) intervals. The scale axis shows divergence times as millions of years ago (MYA).
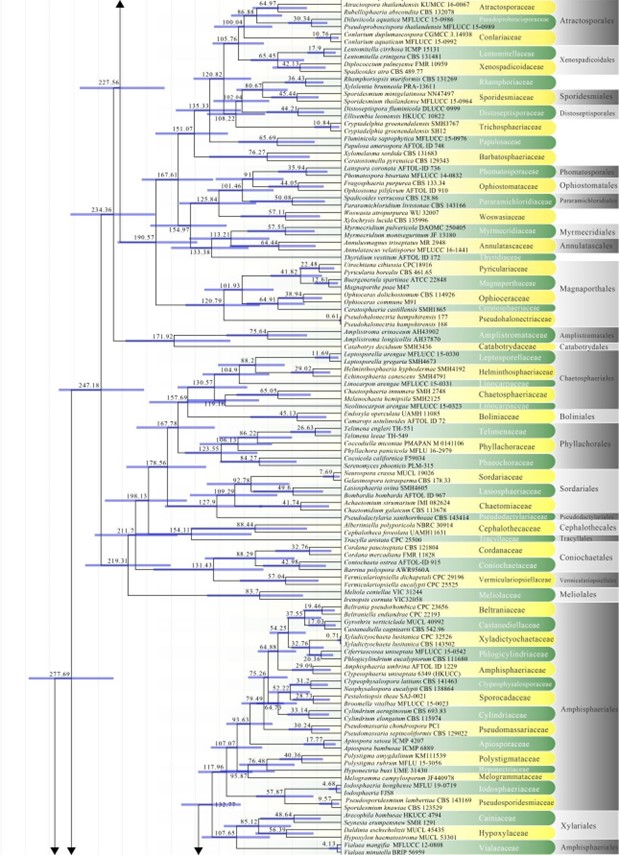
Figure 2 – Continued.
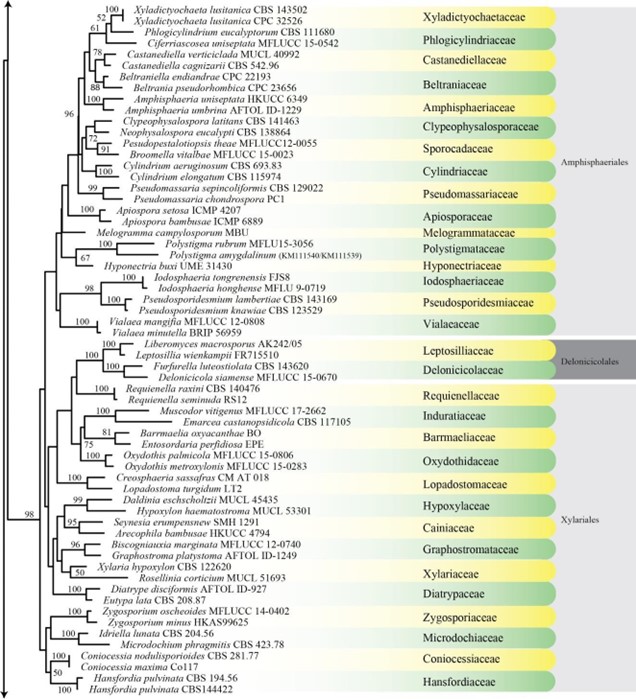
Figure 2 – Continued.
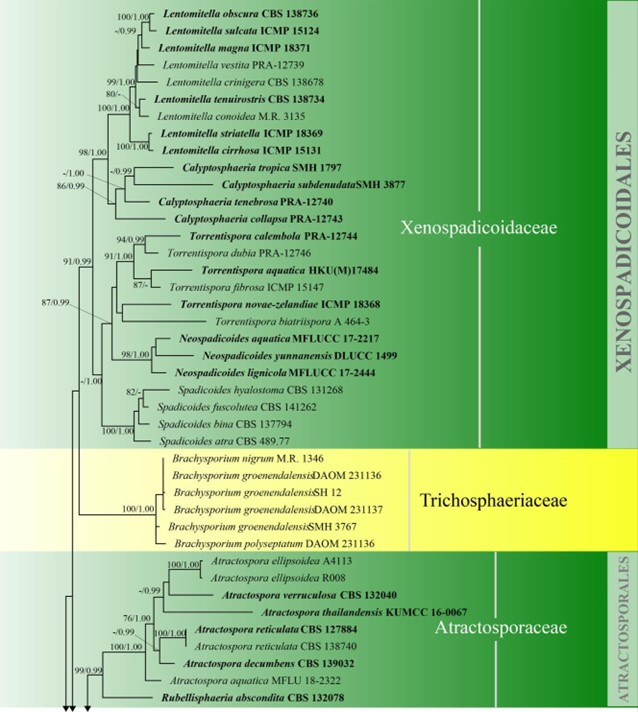
Figure 3 – Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, ITS and rpb2 sequence data of Diaporthomycetidae. One hundred and ninety-three strains are included in the combined analyses which comprised 3545 characters (859 characters for LSU, 972 characters for SSU, 659 characters for ITS) after alignment. Single gene analyses were carried out and the topology of each tree had clade stability. Tree topology of the maximum likelihood analysis is similar to the Bayesian analysis. The best RaxML tree with a final likelihood value of – 68207.368884 is presented. Estimated base frequencies were as follows: A = 0.248206, C = 0.241993, G = 0.285500, T = 0.224301; substitution rates AC = 1.369088, AG = 2.887040, AT = 1.413053, CG = 1.152137, CT = 6.303994, GT = 1.000000; gamma distribution shape parameter a = 0.315782. Bootstrap support values for ML greater than 75% and Bayesian posterior probabilities greater than 0.95 are given near the nodes. The tree is rooted with Diatrype disciformis (AFTOL-ID 927). Ex-type strains are in bold. The newly generated sequences are indicated in blue.
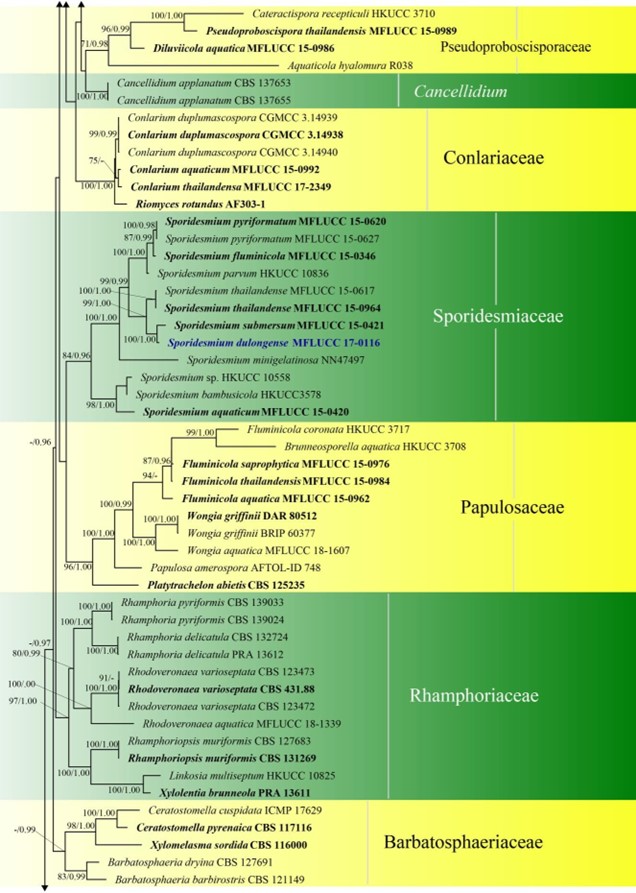
Figure 3 – Continued.
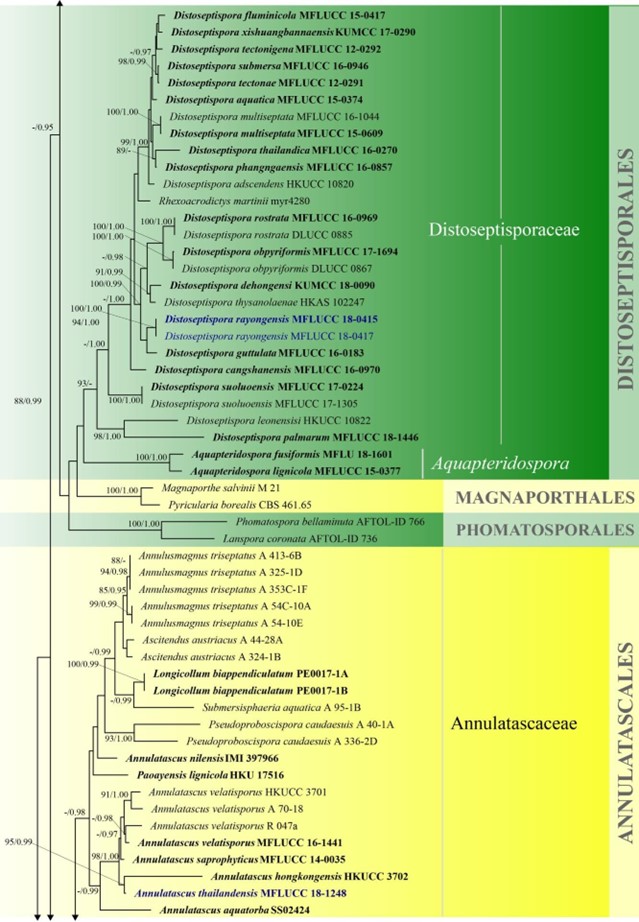
Figure 3 – Continued.
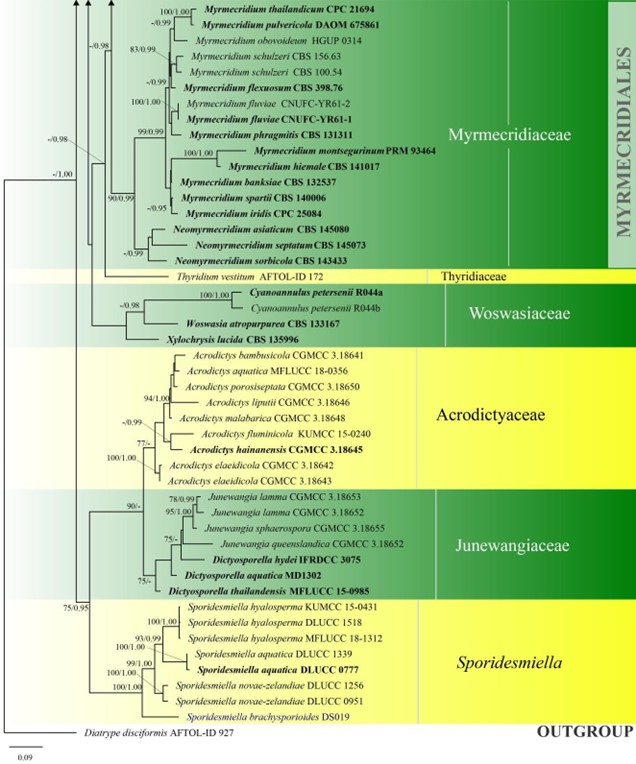
Figure 3 – Continued.
