Ascochyta medicaginicola Q. Chen & L. Cai (2015) Qian Chen & L. Cai, Stud. Mycol. 82: 187 (2015)
= Phoma medicaginis Malbr. & Roum., in Roumeguère, Fungi Selecti Galliaei Exs. 37: no. 3675 (1886)
= Ascochyta medicaginicola var. macrospora Qian Chen & L. Cai, Stud. Mycol. 82: 187 (2015)
= Ascochyta medicaginicola var. medicaginicola Qian Chen & L. Cai, Stud. Mycol. 82: 187 (2015)
Index Fungorum number: IF 814129; MycoBank number: MB 814129; Facesoffungi number: FoF 00423;
Saprobic, endophytic or pathogenic on the host plant in terrestrial habitat (Ellwood et al. 2006; Aveskamp et al. 2008, 2010; Rivera-Orduña et al. 2011). Sexual morph: see Tibpromma et al. (2017). Asexual morph: Conidiomata 150–250 μm diam., 100–150 μm high, dark brown, pycnidial, solitary to gregarious, immersed to semi-immersed, globose, unilocular, thick-walled, ostiolate. Ostiole circular, papillate, centrally located. Conidiomatal wall 15–30 μm wide, composed of thick-walled, dark brown to hyaline cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 4–7 × 5–8 μm, hyaline, enteroblastic, phialidic, doliiform, determinate, smooth-walled. Conidia 10–20 × 3.5–5.5 μm ( ̄x = 15.5 × 5, n = 20), ellipsoidal to ovoid,hyaline, straight or slightly curved, 0–2-septate, constricted at septa, obtuse at both ends, smooth-walled, guttulate.
Material examined – Italy, Province of Ravenna, Zattaglia, on dead twig of Scabiosa sp. (Caprifoliaceae), 30 December 2012, E. Camporesi, IT988 (MFLU 14-0812), living culture, MFLUCC 13-0485, (KUN, HKAS 83971).
Notes – Phoma medicaginis was synonymized under A. medicaginicola var. macrospora by Chen et al. (2015), but this variety together with A. medicaginicola var. medicaginicola are listed as synonyms of Ascochyta medicaginicola in Index Fungorum (2018). We provide a detailed description and photo plate for A. medicaginicola.
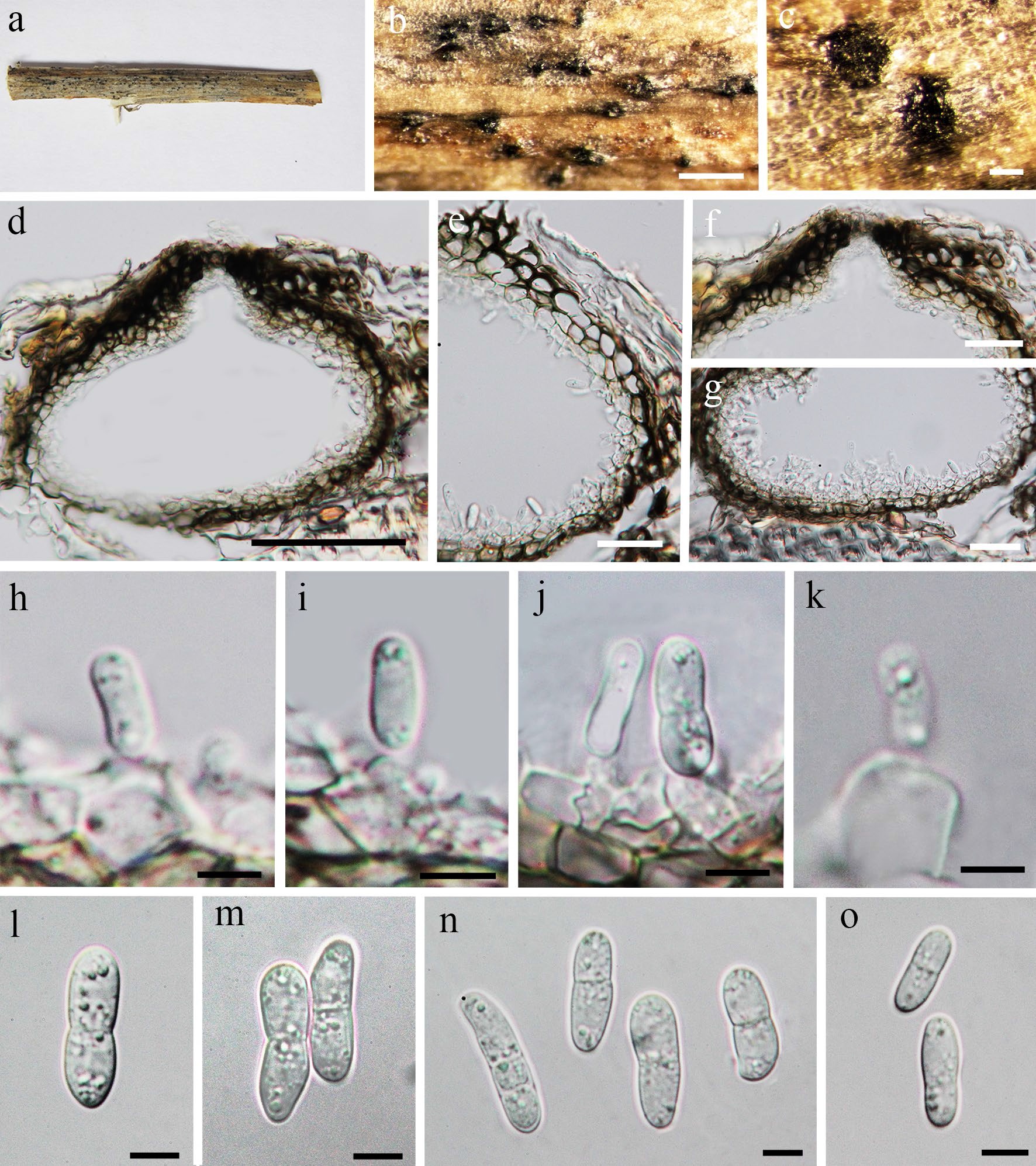
Fig. 1. Ascochyta medicaginicola var. medicaginicola. a, b, c Conidia observed on host substrate. d, e Conidiomata. f, g, h Conidia i, k, l Conidia j Conidiodenous cell. Scale bars: a = 500 µm, b, c, d, e, g = 100 µm, f,h = 50 µm, i, j, k, l = 10 µm.
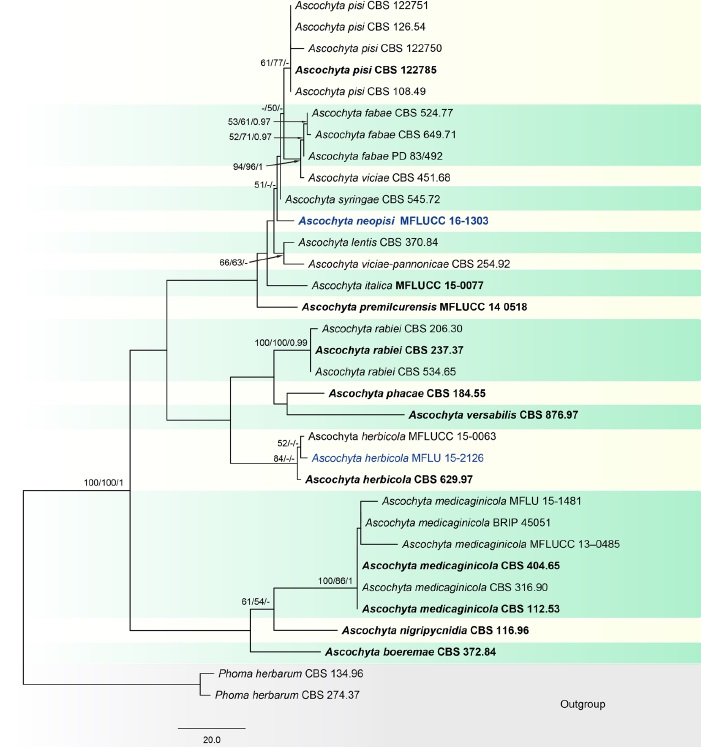
Fig. 1 Phylogenetic tree generated from a maximum parsimony analysis based on a concatenated alignment of ITS, LSU, rpb2 and tub2 sequences data representing Ascochyta. The newly generated nucleotide sequences were compared against the GenBank database by blast search. Related sequences were obtained from Chen et al. (2015), Tibpromma et al. (2017) and GenBank. Thirty-three strains are included in the analyses, which comprise 2273 characters including gaps (ITS: 1–489, LSU: 490–1344, rpb2: 1345–1940, tub2: 1941–2273). Phoma herbarum CBS 134.96 and P. herbarum CBS 274.37 are used as the outgroup taxa. The tree topology of the maximum likelihood analysis is similar to either the maximum parsimony or the Bayesian analysis. The best-scoring RAxML tree with a final optimization likelihood value of − 5657.513625 is presented. The matrix had 270 distinct alignment patterns, with 16.63% of undetermined characters or gaps. Estimated base frequencies were: A = 0.242420, C = 0.238519, G = 0.274514, T = 0.244547; substitution rates AC = 0.891880,
AG = 4.215502, AT = 1.884133, CG = 0.651574, CT = 11.712755, GT = 1.000000; gamma distribution shape parameter α = 0.020000. The maximum parsimonious dataset consisted of constant 1979, 216 parsimony-informative and 78 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 439 steps (CI = 0.749, RI = 0.864, RC = 0.647, HI = 0.251) in the first tree. Maximum parsimony (MPBS) and maximum likelihood (MLBS) bootstrap support values higher than 50%, and Bayesian posterior probabilities ≥ 0.95 (PP) are shown above or below the nodes, respectively. Hyphen (“–”) indicates a value lower than 50% for MPBS and MLBS and a posterior probability lower than 0.95 for BYY. The scale bar indicates 20.0 changes. Ex-type and reference strains are in bold.
